Mecp2 fine-tunes quiescence exit by targeting nuclear receptors
Figures
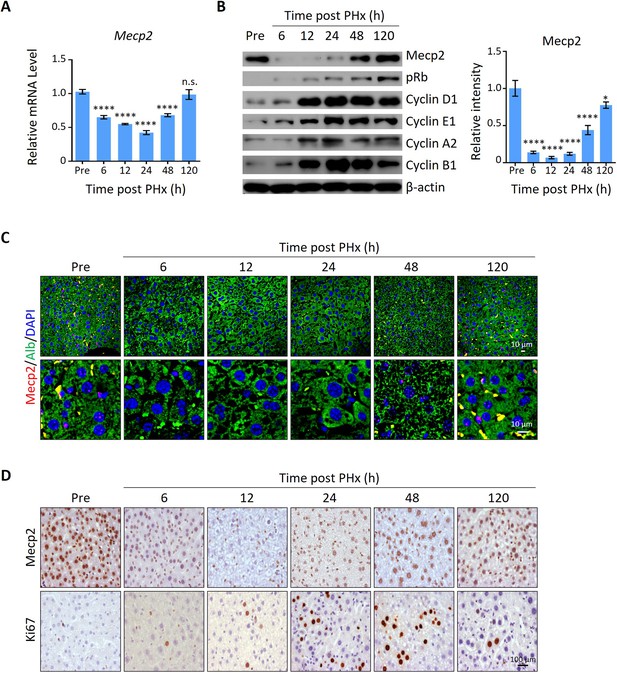
Mecp2 is immediately decreased in the liver after partial hepatectomy (PHx).
(A) Real-time PCR to evaluate mRNA levels of Mecp2 at different time points after PHx. Data are presented as means ± SEM; n = 6. n.s., not significant; ****p<0.0001 by one-way ANOVA. (B) Western blotting (WB) showing the time course of protein levels of Mecp2, pRb, Cyclin D1, Cyclin E1, Cyclin A1, Cyclin B1, and β-actin in mouse livers after PHx. Right panel: quantification of Mecp2. Data are presented as means ± SEM; n = 3. n.s., not significant; ****p<0.0001; *p<0.05 by one-way ANOVA. (C) Representative immunofluorescence (IF) staining of Mecp2 (red) and Alb (green), together with DAPI (blue) for nuclei in liver sections at different time points after PHx. Lower panels: higher-magnification images. (D) Representative immunohistochemistry (IHC) images of liver tissues stained for Mecp2 or Ki67 at the indicated time points after PHx.
-
Figure 1—source data 1
Mecp2 is immediately decreased in the liver after partial hepatectomy (PHx).
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig1-data1-v1.xlsx
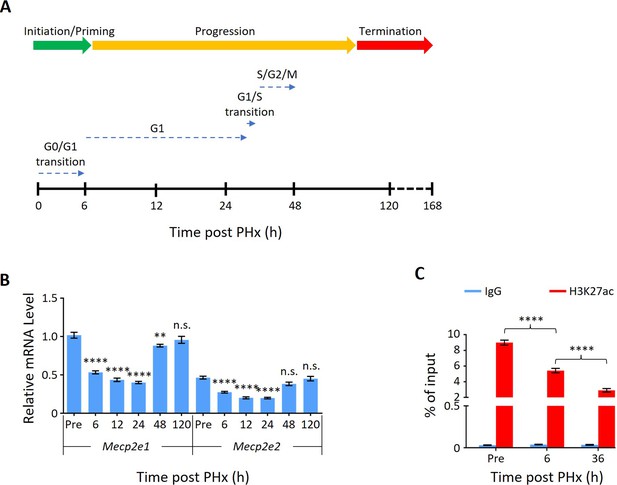
Mecp2 is dynamically expressed during partial hepatectomy (PHx)-induced liver regeneration.
(A) Schematic for the PHx time course and data collection. Three phases of liver regeneration and the cell cycle progression of the first round of cell division after PHx are indicated by rainbow-colored and dotted arrows, respectively. (B) Real-time PCR to evaluate mRNA levels of two isoforms of Mecp2, Mecp2e1 and Mecp2e2, at different time points after PHx. Data are presented as means ± SEM; n = 6; n.s., not significant; **p<0.01; ****p<0.0001 by one-way ANOVA. (C) ChIP-qPCR showing H3K27ac occupancy at the Mecp2 promoter at the indicated time points after PHx. IgG served as a negative control. Data are presented as means ± SEM; n = 6; ****p<0.0001 by two-way ANOVA.
-
Figure 1—figure supplement 1—source data 1
Mecp2 is dynamically expressed during partial hepatectomy (PHx)-induced liver regeneration.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig1-figsupp1-data1-v1.xlsx
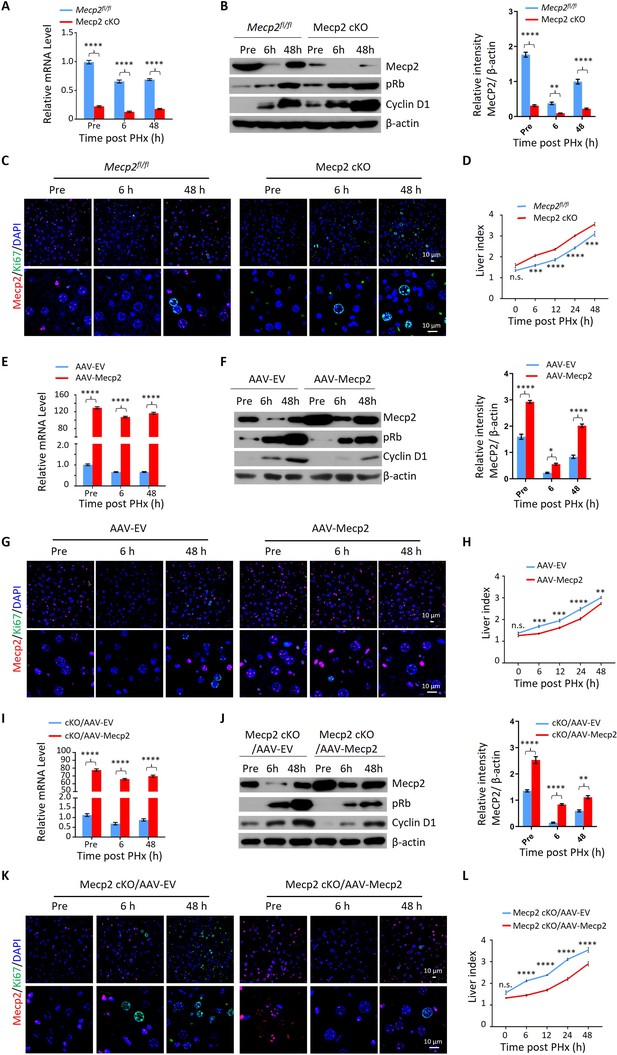
Mecp2 fine-tunes quiescence exit in hepatocytes after partial hepatectomy (PHx) in vivo.
(A–D) Liver regeneration in Mecpfl/fl and Mecp2 cKO mice after PHx. (A) Real-time PCR to measure mRNA levels of Mecp2. The effects and corresponding quantification of Mecp2 KO on quiescence exit and liver regeneration were assessed by western blotting (WB) of Mecp2, pRb, and Cyclin D1 (B), immunofluorescence (IF) staining of Mecp2 (red) and Ki67 (green) in liver sections (C), and liver index of control and Mecp2 cKO mice (D) at the indicated time points. (E–H) Liver regeneration in Mecp2fl/fl livers without (AAV-EV) or with AAV-mediated Mecp2 OE (AAV-Mecp2) after PHx. AAV, adeno-associated virus; EV, empty vector. (E) Real-time PCR to measure mRNA levels of Mecp2. (F–H) The effects and corresponding quantification of Mecp2 OE on quiescence exit and liver regeneration were assessed by WB of Mecp2, pRb, and Cyclin D1 (F), IF staining of Mecp2 (red) and Ki67 (green) in liver sections (G), and liver index at the indicated time points (H). (I–L) Liver regeneration in Mecp2 cKO livers without (Mecp2 cKO/AAV-EV) or with AAV-mediated Mecp2 restoration (Mecp2 cKO/AAV-Mecp2) after PHx. (I) Real-time PCR to measure mRNA levels of Mecp2. (J–L) The effects and corresponding quantification of Mecp2 restoration on quiescence exit and liver regeneration in Mecp2 cKO livers were assessed by WB of Mecp2, pRb and Cyclin D1 (J), IF staining of Mecp2 (red) and Ki67 (green) in liver sections (K), and liver index (L) at the indicated time points. Data are presented as means ± SEM. In (A, E, I), n = 6; (B, F, J), n = 3; in (D, H, L), n = 5 mice/group. n.s., not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 by two-way ANOVA.
-
Figure 2—source data 1
Mecp2 fine-tunes quiescence exit in hepatocytes after partial hepatectomy (PHx) in vivo.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig2-data1-v1.xlsx
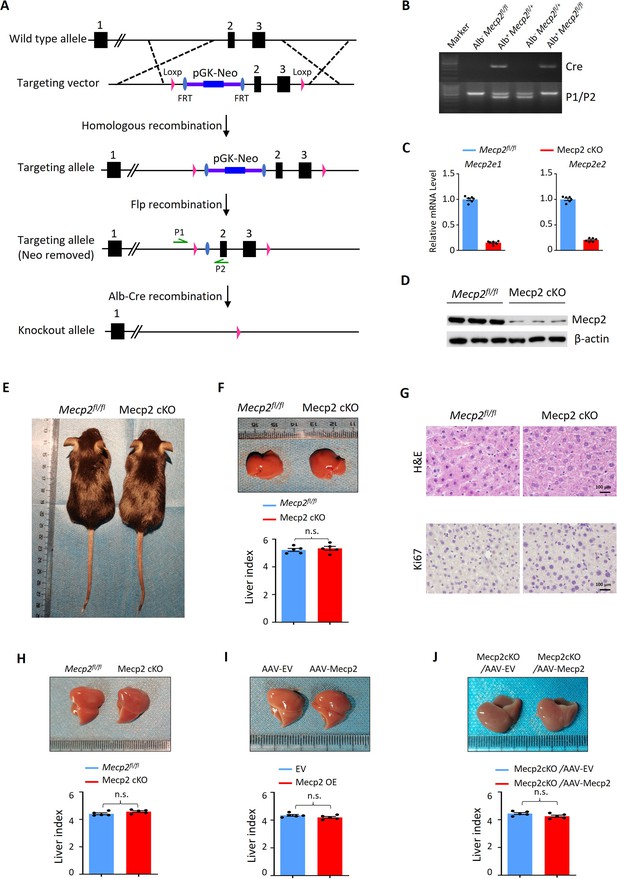
Modulation of Mecp2 expression in the mouse liver.
(A) Schematic diagram of loss of Mecp2 in mouse hepatocyte. In the Mecp2 floxed allele, Mecp2 exons 2 and 3 were flanked with loxp sites. The Neo-cassette was removed by mating with FLP recombinase homozygous mice. Exons 2 and 3 were deleted by mating with Alb-Cre recombinase homozygous mice. (B–D) Evaluation of hepatocyte-specific depletion of Mecp2 by genotyping (B), real-time PCR (C), and western blotting (WB) (D) in Mecp2fl/fl and Mecp2 cKO livers. Data are presented as means ± SEM. n = 6 mice/group; ****p<0.0001 by one-way ANOVA. (E) Representative images of Mecp2fl/fl and Mecp2 cKO mice at 3 months of age. (F) Representative liver morphology and liver index of Mecp2fl/fl and Mecp2 cKO mice before PHx. Data are presented as means ± SEM. n = 5 mice/group; n.s., not significant by Student’s t-test. (G) Representative H&E staining and immunohistochemistry (IHC) for Ki67 images of liver tissues from Mecp2fl/fl and Mecp2 cKO mice before PHx. (H) Representative liver morphology and liver index of Mecp2fl/fl and Mecp2 cKO mice at 7 d post-PHx. Data are presented as means ± SEM. n = 5 mice/group; n.s., not significant by Student’s t-test. (I) Representative liver morphology and liver index of Mecp2fl/fl mice without (AAV-EV) or with (AAV-Mecp2) AAV-mediated Mecp2 overexpression at 7 d post-PHx. EV, empty vector. Data are presented as means ± SEM. n = 5 mice/group; n.s., not significant by Student’s t-test. (J) Representative liver morphology and liver index of Mecp2-cKO mice without (Mecp2 cKO/AAV-EV) or with (Mecp2 cKO/AAV-Mecp2) the rescue of Mecp2 at 7 d post-PHx. Data are presented as means ± SEM. n = 5 mice/group; n.s., not significant by Student’s t-test.
-
Figure 2—figure supplement 1—source data 1
Modulation of Mecp2 expression in the mouse liver.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig2-figsupp1-data1-v1.xlsx
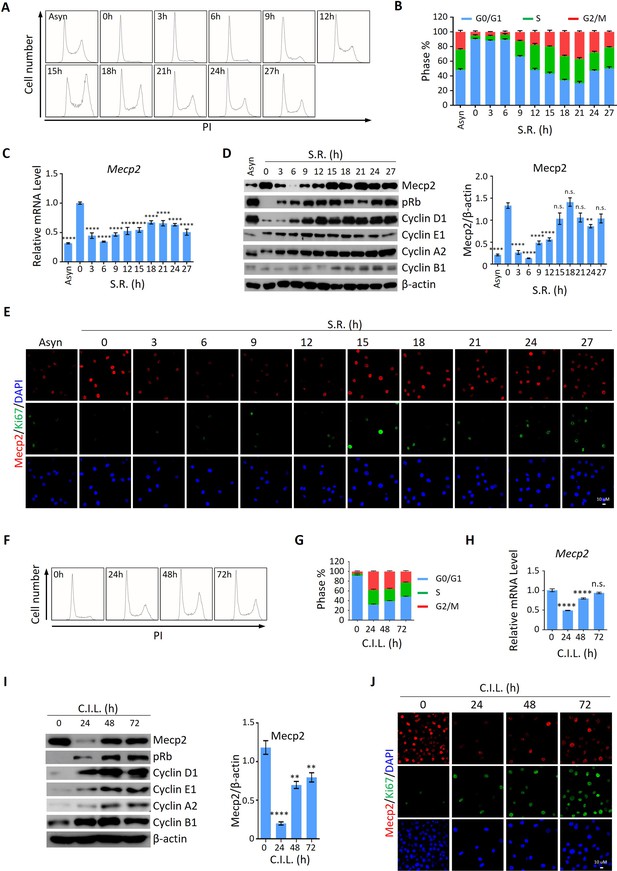
Mecp2 is immediately decreased during quiescence exit in cellular models.
(A, B) Representative histograms of propidium iodide (PI) staining of either asynchronized (Asyn) proliferating or starvation-induced quiescent 3T3 cells after serum restimulation (SR) in (A) and statistical analysis of cell cycle distribution (B). (C) Real-time PCR to examine mRNA levels of Mecp2. Data are presented as means ± SEM; n = 9. n.s., not significant; ****p<0.0001 by one-way ANOVA. (D) Western blotting (WB) of Mecp2, pRb, Cyclin D1, Cyclin E1, Cyclin A1, and Cyclin B1 in quiescent 3T3 cells upon SR. Right panel: quantification of Mecp2. Data are presented as means ± SEM; n = 3. n.s., not significant; ****p<0.0001 by one-way ANOVA. (E) Representative immunofluorescence (IF) staining of Mecp2 (red) and Ki67 (green), together with DAPI (blue) in Asyn and quiescent 3T3 cells upon SR. (F, G) Representative histograms of PI staining of contact inhibition (CI)-induced quiescent 3T3 cells after CI loss (CIL) (F) and statistical analysis of cell cycle distribution (G) at the indicated time points. (H) Real-time PCR to examine mRNA levels of Mecp2 in 3T3 cells released from CI-induced quiescence. Data are presented as means ± SEM; n = 6. n.s., not significant; ****p<0.0001 by one-way ANOVA. (I) WB of Mecp2, pRb, Cyclin D1, Cyclin E1, Cyclin A1, and Cyclin B1 in quiescent 3T3 cells upon CIL. Right panel: quantification of Mecp2. Data are presented as means ± SEM; n = 3. n.s., not significant; ****p<0.0001 by one-way ANOVA. (J) Representative IF staining of Mecp2 and Ki67 in 3T3 cells released from CI-induced quiescence.
-
Figure 3—source data 1
Mecp2 is immediately decreased during quiescence exit in cellular models.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig3-data1-v1.xlsx
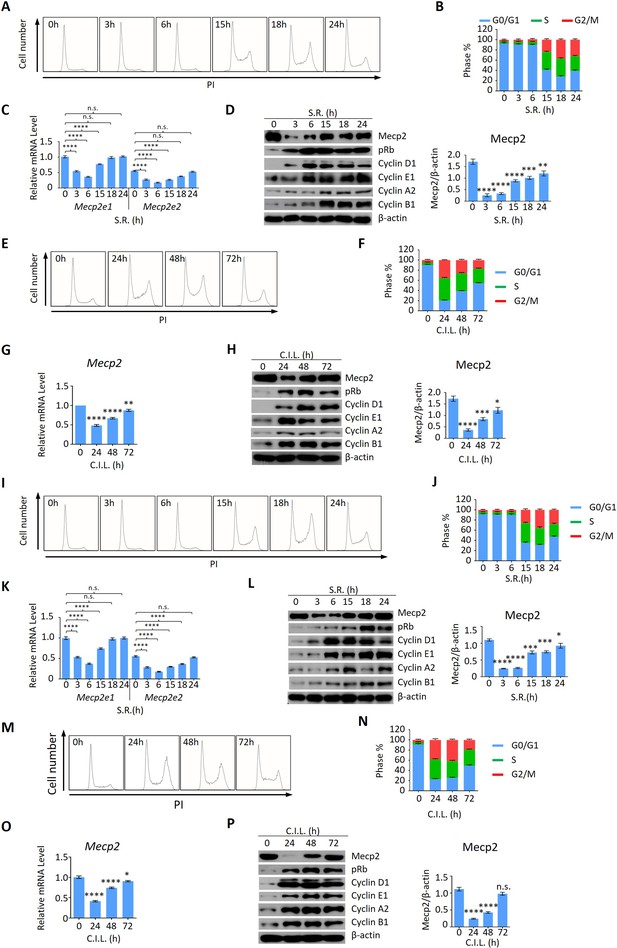
Mecp2 is dynamically expressed during quiescence exit in both HT22 and human umbilical vein endothelial cells (HUVECs).
Serum restimulation (SR)-induced (A–D, I–L) or contact inhibition loss (CIL)-induced (E–H, M–P) quiescence exits in mouse hippocampal neuronal HT22 cells (A–H) or in HUVECs (I–P) was analyzed by propidium iodide (PI) staining (A, B, E, F, I, J, M, N), real-time PCR (C, G, K, O), and western blotting (WB) (D, H, L, P) in a time course of quiescence exit. (A, E, I, M) Representative histograms of PI staining. (B, F, J, N) Statistical analysis of the cell cycle distribution. (D, H, L, P) WB of Mecp2, pRb, Cyclin D1, Cyclin E1, Cyclin A1, Cyclin B1, and β-actin in cells released from G0. Left panel: quantification of Mecp2 protein levels. In (C, K), n = 9; In (O, P), n = 6; In (D, G, H, L) n = 3; Data are presented as means ± SEM; n.s., not significant; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 by one-way ANOVA.
-
Figure 3—figure supplement 1—source data 1
Mecp2 is dynamically expressed during quiescence exit in both HT22 and human umbilical vein endothelial cells (HUVECs).
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig3-figsupp1-data1-v1.xlsx
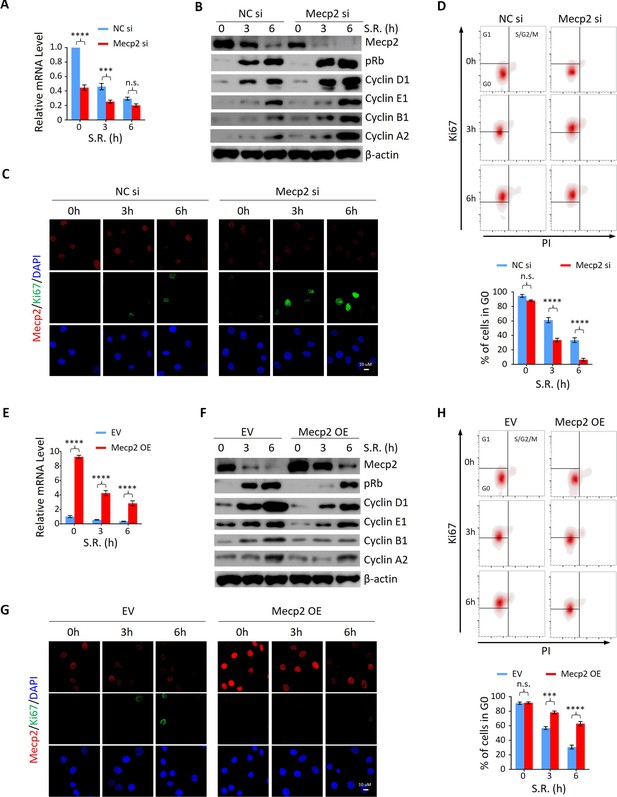
Mecp2 negatively regulates the G0/G1 transition in the cellular model of serum restimulation (SR)-induced quiescence exit.
(A) Real-time PCR showing the mRNA levels of Mecp2 in 3T3 cells transfected with negative control siRNA (NC si) or Mecp2 siRNA (Mecp2 si) at the early stages of SR-induced cell cycle reentry. Data are presented as means ± SEM; n = 6. n.s., not significant; ***p<0.001; ****p<0.0001 by two-way ANOVA. (B) Western blotting (WB) of Mecp2, pRb, Cyclin D1, Cyclin E1, Cyclin A2, and Cyclin B1 in control and Mecp2 knockdown (KD) 3T3 cells released from serum starvation (SS)-induced quiescence at the indicated time points. (C) Representative immunofluorescence (IF) staining of Mecp2 and Ki67 in control and Mecp2 KD 3T3 cells upon SR-induced quiescent exit. (D) Ki67 and propidium iodide (PI) double staining followed by flow cytometry showing cell cycle profiles of 3T3 cells transfected with NC or Mecp2 siRNA upon SR-induced quiescence exit. Cells in G0, G1, and S/G2/M phases were defined by Ki67−/2N DNA content, Ki67+/2N DNA content and >2N DNA content population, respectively. Lower panel: quantification of the percentage of 3T3 cells in the G0 phase. Data are presented as means ± SEM; n = 3. n.s., not significant; ****p<0.0001 by two-way ANOVA. (E) Real-time PCR showing the mRNA levels of Mecp2 in 3T3 cells transduced with the empty vector (EV) or the vector overexpressing Mecp2 (Mecp2 overexpression [OE]) at the early stages of quiescence exit. Data are presented as means ± SEM; n = 3. ****p<0.0001 by two-way ANOVA. (F) WB of Mecp2, pRb, Cyclin D1, Cyclin E1, Cyclin A2, and Cyclin B1 in control and Mecp2 OE 3T3 cells released from SS-induced quiescence at the indicated time points. (G) Representative IF staining of Mecp2 and Ki67 in quiescent control and Mecp2 OE 3T3 cells upon SR. (H) Representative flow cytometry plots of Ki67/PI double staining in control and Mecp2 OE 3T3 cells upon SR-induced quiescence exit. Lower panel: quantification of proportion of 3T3 cells in the G0 phase. Data are presented as means ± SEM; n = 3. n.s., not significant; ***p<0.001; ****p<0.0001 by two-way ANOVA.
-
Figure 4—source data 1
Mecp2 negatively regulates the G0/G1 transition in the cellular model of serum restimulation (SR)-induced quiescence exit.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig4-data1-v1.xlsx
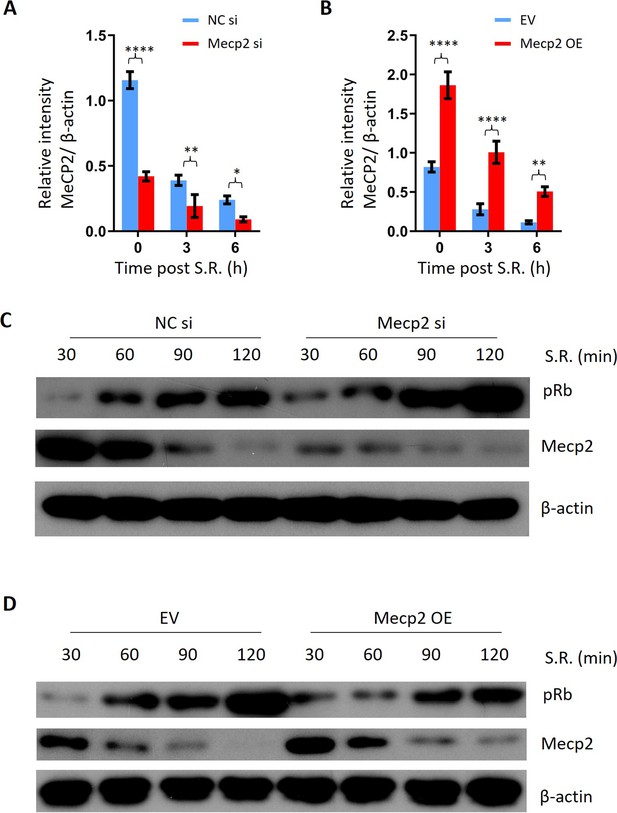
Mecp2 negatively regulates the G0/G1 transition in the cellular model of serum restimulation (SR)-induced quiescence exit.
(A) Protein quantitation of MeCP2 protein expression normalized to β-actin for Figure 4B western blotting (WB). Data are presented as means ± SEM; n = 3. *p<0.05; ***p<0.001; ****p<0.0001 by two-way ANOVA. (B) Protein quantitation of MeCP2 protein expression normalized to β-actin for Figure 4F western blotting. Data are presented as means ± SEM; n = 3. **p<0.01; ****p<0.0001 by two-way ANOVA. (C) WB of Mecp2 and pRb in control and Mecp2 knockdown (KD) 3T3 cells released from serum starvation (SS)-induced quiescence at the indicated time points. (D) WB of Mecp2 and pRb in control and Mecp2 overexpression (OE) 3T3 cells released from SS-induced quiescence at the indicated time points.
-
Figure 4—figure supplement 1—source data 1
Mecp2 negatively regulates the G0/G1 transition in the cellular model of serum restimulation (SR)-induced quiescence exit.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig4-figsupp1-data1-v1.xlsx
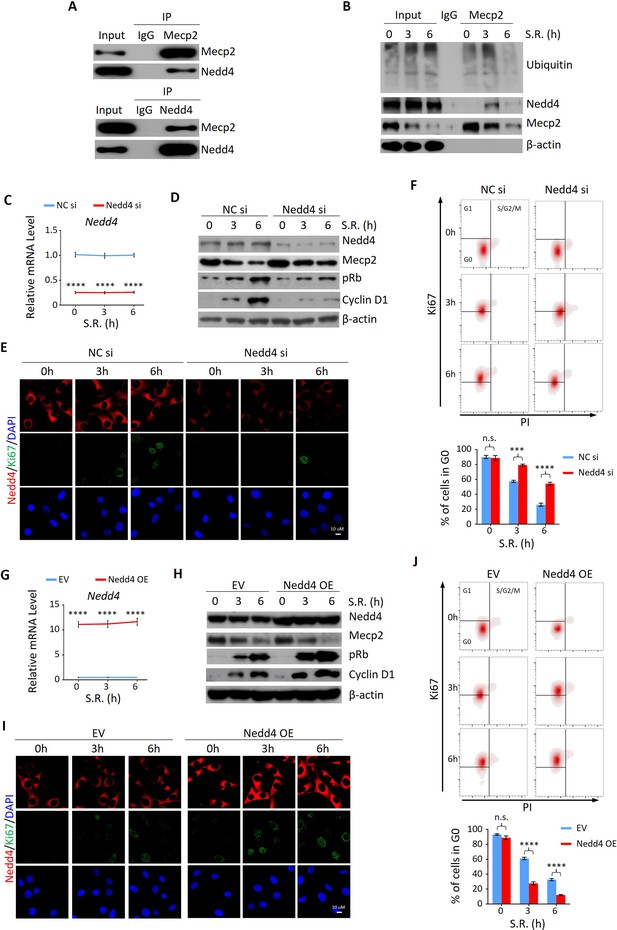
Nedd4 interacts with Mecp2 and affects quiescence exit by facilitating Mecp2 degradation.
(A) Reciprocal immunoprecipitation-western blotting (IP-WB) analysis to validate the interaction between endogenous Mecp2 and Nedd4. (B) Co-IP of Mecp2, ubiquitin, and Nedd4 in quiescent 3T3 cells during serum restimulation (SR)-induced quiescence exit. (C) Real-time PCR showing siRNA-mediated Nedd4 knockdown (KD) in 3T3 cells upon SR-induced quiescence exit. Data are presented as means ± SEM; n = 5. ****p<0.0001 by two-way ANOVA. (D–F) The effect of Nedd4 KD on quiescent exit in 3T3 cells determined by WB (D), immunofluorescence (IF) staining of Ki67 and Nedd4 (E), and Ki67/PI staining followed by flow cytometry (F) at the indicated time points. Lower panel in (F): quantification of the percentage of 3T3 cells in the G0 phase. Data are presented as means ± SEM; n = 3. n.s., not significant; ***p<0.001, ****p<0.0001 by two-way ANOVA. (G) Real-time PCR showing Nedd4 overexpression (OE) in 3T3 cells upon SR-induced quiescence exit. Data are presented as means ± SEM; n = 5. ****p<0.0001 by two-way ANOVA. (H–J) The effect of Nedd4 OE on quiescent exit in 3T3 cells determined by WB (H), IF staining of Ki67 and Nedd4 (I), and Ki67/PI staining (J) at the indicated time points. Data are presented as means ± SEM; n = 3. n.s., not significant; ****p<0.0001 by two-way ANOVA.
-
Figure 5—source data 1
Nedd4 interacts with Mecp2 and affects quiescence exit by facilitating Mecp2 degradation.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig5-data1-v1.xlsx
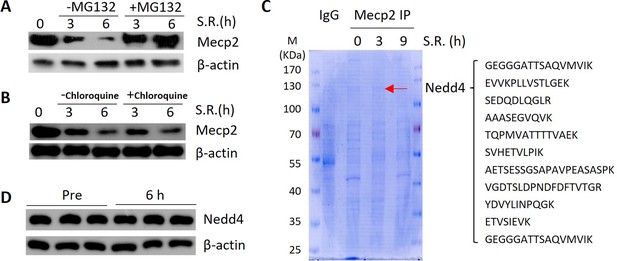
Nedd4 interacts with Mecp2.
(A) Western blotting (WB) of Mecp2 in quiescent 3T3 cells treated with or without MG132 after being released from G0 by serum restimulation (SR). (B) WB of Mecp2 in quiescent 3T3 cells treated with or without chloroquine after being released from G0 by SR. (C) Coomassie staining of Mecp2 co-IP proteins separated by SDS-PAGE. Right: representative peptides of Nedd4 identified by mass spectrometry. (D) WB showing protein levels of Nedd4 in mouse livers before and 6 hr after partial hepatectomy (PHx).
-
Figure 5—figure supplement 1—source data 1
Nedd4 interacts with Mecp2.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig5-figsupp1-data1-v1.xlsx
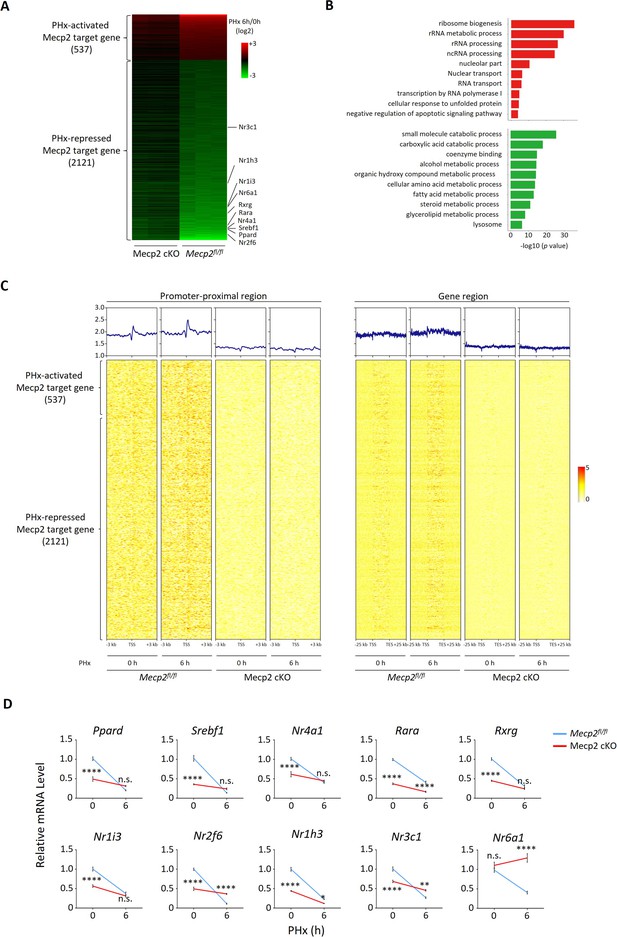
Mecp2 transcriptionally regulates quiescence exit.
(A) Heatmap of Mecp2 direct target genes at the early stage of liver regeneration rank-ordered by their gene expression fold change. (B) The top 10 most significantly overrepresented Gene Ontology (GO) terms for the partial hepatectomy (PHx)-activated (red) and PHx-repressed (green) Mecp2 target genes. (C) Heatmaps depicting ChIP-seq enrichment of Mecp2 at the promoter-proximal region (3 kb away from transcription start sites [TSS]) and the defined gene region of Mecp2 target genes in Mecpfl/fl and Mecp2 cKO livers before and 6 hr post-PHx. Genes are rank-ordered according to the fold change of expression. (D) Real-time PCR validation of PHx-repressed NRs in Mecp2fl/fl and Mecp2 cKO livers upon PHx. Data are presented as means ± SEM; n = 5. n.s., not significant; **p<0.01; ****p<0.0001 by two-way ANOVA.
-
Figure 6—source data 1
Mecp2 transcriptionally regulates quiescence exit.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig6-data1-v1.xlsx
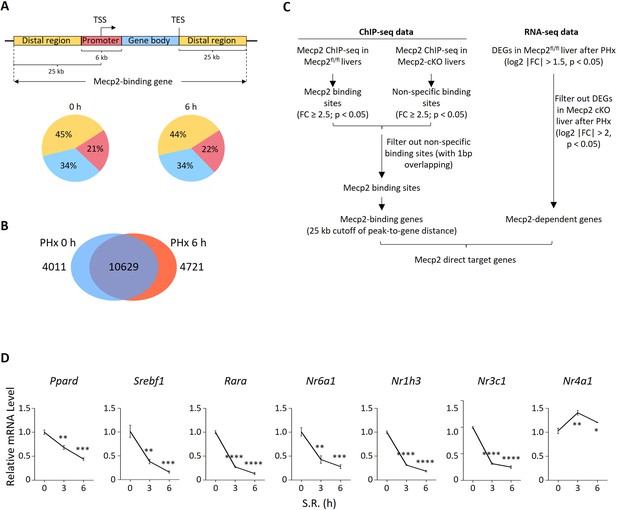
Related to Figure 6.
(A) Schematic showing the definition of Mecp2-binding genes. Lower panel: distribution of Mecp2 peaks in defined promoter-proximal, gene body, and distal regions. TSS, transcriptional start site; TES, transcriptional end site. The cutoff of distance of peak-to-gene is 25 kb. (B) Venn diagram overlap of Mecp2 binding genes in Mecp2fl/fl livers before (0 hr) and after partial hepatectomy (PHx) (6 hr). (C) Pipeline showing the integration of Mecp2 ChIP-seq data with RNA-seq data to identify Mecp2-direct target genes. (D) Real-time PCR validation of PHx-repressed NRs in 3T3 cells upon serum restimulation (SR)-induced quiescence exit. Data are presented as means ± SEM; n = 3. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 by one-way ANOVA.
-
Figure 6—figure supplement 1—source data 1
Raw data related to Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig6-figsupp1-data1-v1.xlsx
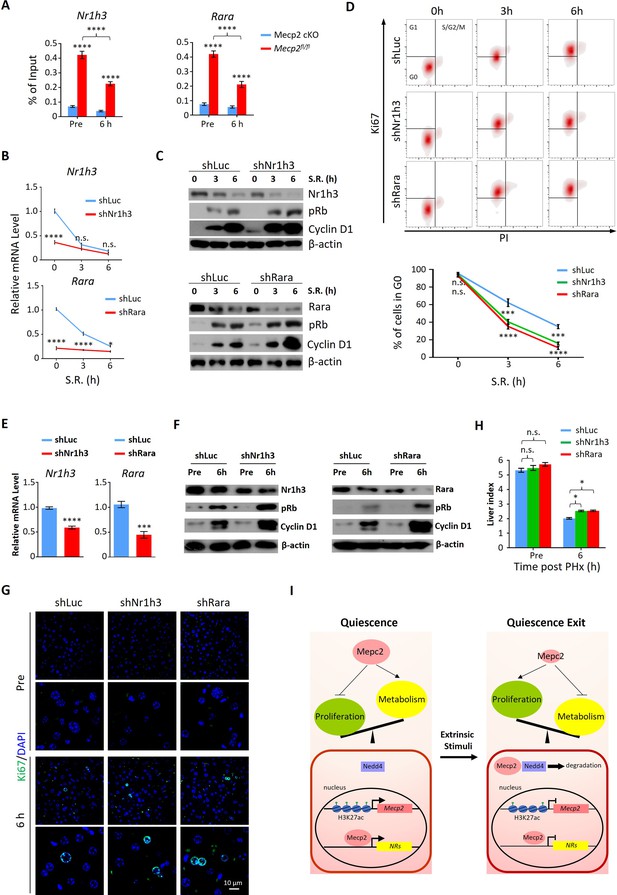
Depletion of either Nr1h3 or Rara mimics the Mecp2 knockdown (KD) phenotype during quiescence exit.
(A) ChIP-qPCR analyses of Mecp2 at the promoter-proximal regions of Rara and Nr1h3 in Mecp2fl/fl and Mecp2-cKO livers before and 6 hr post-partial hepatectomy (PHx). (B–D) Either Nr1h3 or Rara KD promotes serum restimulation (SR)-induced quiescence exit in 3T3 cells. (B) Real-time PCR showing lentivirus-mediated KD of either Nr1h3 or Rara. shLuc served as a negative control. (C) Western blotting (WB) of pRb, Cyclin D1, Nr1h3, and Rara in control and Nr1h3 or Rara KD 3T3 cells at the indicated time points. (D) The effect of Nr1h3 or Rara KD on quiescent exit in 3T3 cells determined by Ki67/PI staining followed by flow cytometry. Data are presented as means ± SEM; In (A, B), n = 5; in (D), n = 3. *p<0.05; ***p<0.001; ****p<0.0001 by two-way ANOVA. (E–H) Either Nr1h3 or Rara KD further enhances quiescence exit in Mecp2 cKO livers. (E) Real-time PCR showing adeno-associated virus (AAV)-mediated KD of either Nr1h3 or Rara. shLuc served as a negative control. (F) WB of pRb, Cyclin D1, Nr1h3, and Rara in control and Nr1h3 or Rara KD 3T3 cells at the indicated time points. (G) The effect of Nr1h3 or Rara KD on quiescent exit in Mecp2 cKO livers determined by IF and liver index (H) before and 6 hr post-PHx. Data are presented as means ± SEM. In (E, H), n = 5 mice/group; n.s., not significant; *p<0.05, ****p<0.0001 by two-way ANOVA. (I) Model of the negative regulatory role for Mecp2 in fine-tuning quiescence exit.
-
Figure 7—source data 1
Depletion of either Nr1h3 or Rara mimics the Mecp2 KD phenotype during quiescence exit.
- https://cdn.elifesciences.org/articles/89912/elife-89912-fig7-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HUVEC | ATCC | Cat# CRL-1730 | |
| Cell line (H. sapiens) | LentiX-293T | ATCC | Cat# ACS-4500 | |
| Cell line (Mus musculus) | NIH3T3 | ATCC | Cat# SCSP-515 | |
| Cell line (M. musculus) | Ht22 | Procell Life Science&Technology | Cat# CL-0697 | |
| Antibody | Albumin (mouse monoclonal) | Proteintech | Cat# 66051-1-Ig; RRID:AB_11042320 | 1:100 |
| Antibody | β-Actin (mouse monoclonal) | Proteintech | Cat# 66009-1-Ig; RRID:AB_2687938 | 1:4000 |
| Antibody | Cyclin A2 (mouse monoclonal) | abcam | Cat# ab38; RRID:AB_304084 | 1:1000 |
| Antibody | Cyclin B1 (mouse monoclonal) | abcam | Cat# ab72; RRID:AB_305751 | 1:1000 |
| Antibody | Cyclin D1 (rabbit monoclonal) | abcam | Cat# ab16663; RRID:AB_443423 | 1:1000 |
| Antibody | Cyclin E1 (rabbit monoclonal) | Cell Signaling Technology | Cat# 20808; RRID:AB_2783554 | 1:1000 |
| Antibody | H3K27ac (rabbit polyclonal) | abcam | Cat# ab4729; RRID:AB_2118291 | 1:50 |
| Antibody | IgG (rabbit, IgG) | Cell Signaling Technology | Cat# 2729; RRID:AB_1031062 | 1:50 |
| Antibody | K48-ubiquitin (rabbit monoclonal) | abcam | Cat# ab140601 | 1:1000 |
| Antibody | Ki67-Immunofluorescence (mouse monoclonal) | abcam | Cat# ab279653 | 1:100 |
| Antibody | Ki67-FACS (rat monoclonal) | BioLegend | Cat# 652406; RRID:AB_2561930 | 1:100 |
| Antibody | MeCP2 (rabbit monoclonal) | Cell Signaling Technology | Cat# 3456; RRID:AB_2143849 | 1:1000 |
| Antibody | MeCP2-ChIP (rabbit polyclonal) | abcam | Cat# ab2828; RRID:AB_2143853 | 1:50 |
| Antibody | Nedd4 (rabbit polyclonal) | Proteintech | Cat# 21698-1-AP; RRID:AB_10858626 | 1:1000 |
| Antibody | Nr1h3 (rabbit polyclonal) | Proteintech | Cat# 14351-1-AP; RRID:AB_10640525 | 1:1000 |
| Antibody | Rara (rabbit polyclonal) | Proteintech | Cat# 10331-1-AP; RRID:AB_2177742 | 1:1000 |
| Antibody | p-Rb S807/811 (rabbit monoclonal) | Cell Signaling Technology | Cat# 8516; RRID:AB_11178658 | 1:1000 |
| Antibody | Ubiquitin (mouse monoclonal) | Cell Signaling Technology | Cat# 3936; RRID:AB_331292 | 1:1000 |
| Antibody | Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (mouse polyclonal) | Thermo Fisher Scientific | Cat# A21202; RRID:AB_141607 | 1:100 |
| Antibody | Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (rabbit polyclonal) | Thermo Fisher Scientific | Cat# A21207; RRID:AB_141637 | 1:100 |
| Recombinant DNA reagent | pLKO.1 vector | Addgene | Cat #8453; RRID:Addgene_8453 | |
| Recombinant DNA reagent | pCMV-deltaR8 | Addgene | Cat #12263; RRID:Addgene_12263 | |
| Recombinant DNA reagent | pCMV-VsVg | Addgene | Cat #8454; RRID:Addgene_8454 | |
| Recombinant DNA reagent | AAV8-TBG-MeCP2 | Genechem Co., Ltd | GOSV0233517 | |
| Recombinant DNA reagent | Mecp2 Mouse Tagged ORF Clone, transcript variant 1 | OriGene | Cat# MR226839 | |
| Recombinant DNA reagent | Mecp2 Mouse Tagged ORF Clone, transcript variant 2 | OriGene | Cat# MR207745 | |
| Recombinant DNA reagent | Nedd4 Mouse Tagged ORF Clone | OriGene | Cat# MR222243 | |
| Recombinant DNA reagent | pCMV6-Entry Mammalian Expression Vector | OriGene | Cat# PS100001 | |
| Chemical compound, drug | MG132 | selleck | Cat# S2619 | 10 μM |
| Chemical compound, drug | Puromycin | Sigma-Aldrich | Cat# P8833 | 2 µg/ml |
| Chemical compound, drug | Polybrene | Sigma-Aldrich | Cat# TR-1003 | 6 µg/ml |
| Chemical compound, drug | Pen Strep | Gibco | Cat# 15140-122 | 1% |
| Commercial assay or kit | Complete Protease Inhibitor mini EASY packs EDTA-Free | Roche | Cat# 05892791001 | |
| Commercial assay or kit | Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000-015 | |
| Commercial assay or kit | Propidium Iodide (PI)/RNase Staining Solution | Cell Signaling Technology | Cat# 4087 | |
| Commercial assay or kit | DAPI | Thermo Fisher Scientific | Cat# D-1306 | |
| Commercial assay or kit | Pierce IP Lysis Buffer | Thermo Fisher Scientific | Cat# 87787 | |
| Commercial assay or kit | DAB Kit | ZSGB-BIO | Cat# ZLI-9018 | |
| Commercial assay or kit | Western Lightning Plus ECL | PerkinElmer | Cat# 0RT2655 | |
| Commercial assay or kit | SimpleChIP Plus Enzymatic Chromatin IP Kit | Cell Signaling Technology | Cat# 9005 | |
| Commercial assay or kit | Dynabeads Protein G | Thermo Fisher Scientific | Cat# 10004D | |
| Commercial assay or kit | Coomassie Blue Super Fast Staining Solution | Beyotime | Cat# P0017F | |
| Sequence-based reagent | siRNA to MeCP2 #1 | This paper | Suzhou GenePharma Co., Ltd | CCUGAAGGUUGGACACGAA |
| Sequence-based reagent | siRNA to MeCP2 #2 | This paper | Suzhou GenePharma Co., Ltd | UGACAAAGCUUCCCGAUUA |
| Sequence-based reagent | siRNA to MeCP2 #3 | This paper | Suzhou GenePharma Co., Ltd | CCGAAUUGCUGCUGCUUUA |
| Sequence-based reagent | siRNA to MeCP2 #4 | This paper | Suzhou GenePharma Co., Ltd | CGAAAUGGCUGUGUAGCAA |
| Sequence-based reagent | siRNA to Nedd4: #1 | This paper | Suzhou GenePharma Co., Ltd | CAGUGAUCCUUACGUAAGATT |
| Sequence-based reagent | siRNA to Nedd4 #2 | This paper | Suzhou GenePharma Co., Ltd | GGGAAAUCGUACGAGAAGATT |
| Sequence-based reagent | siRNA to Nedd4 #3 | This paper | Suzhou GenePharma Co., Ltd | GGAGGAUUAUGGGUGUGAATT |
| Sequence-based reagent | Alb, F | This paper | PCR primer | ACCTGAAGATGTTCGCGATTATCT |
| Sequence-based reagent | Alb, R | This paper | PCR primer | ACCGTCAGTACGTGAGATATCTT |
| Sequence-based reagent | MeCP2, F | This paper | PCR primer | GCTGGGGCCCTTGTTTTGAAT |
| Sequence-based reagent | MeCP2, R | This paper | PCR primer | GCTTTAGGTTGCTGGTGATA |
| Sequence-based reagent | Ar, F | This paper | PCR primer | CTGGGAAGGGTCTACCCAC |
| Sequence-based reagent | Ar, R | This paper | PCR primer | GGTGCTATGTTAGCGGCCTC |
| Sequence-based reagent | α-actin mouse, F | This paper | PCR primer | GGCTGTATTCCCCTCCATCG |
| Sequence-based reagent | α-actin mouse, R | This paper | PCR primer | CCAGTTGGTAACAATGCCATGT |
| Sequence-based reagent | α-actin human, F | This paper | PCR primer | CACCATTGGCAATGAGCGGTTC |
| Sequence-based reagent | α-actin human, R | This paper | PCR primer | AGGTCTTTGCGGATGTCCACGT |
| Sequence-based reagent | MeCP2 common, F | This paper | PCR primer | TATTTGATCAATCCCCAGGG |
| Sequence-based reagent | MeCP2 common, R | This paper | PCR primer | CTCCCTCTCCCAGTTACCGT |
| Sequence-based reagent | MeCP2 mouse, F | This paper | PCR primer | GAGCGGCACTGGGAGACC |
| Sequence-based reagent | MeCP2 mouse, R | This paper | PCR primer | CTGGATGGTGGTGATGAT |
| Sequence-based reagent | MeCP2 human, F | This paper | PCR primer | GATGTGTATTTGATCAATCCC |
| Sequence-based reagent | MeCP2 human, R | This paper | PCR primer | TTAGGGTCCAGGGATGTGTC |
| Sequence-based reagent | Nedd4, F | This paper | PCR primer | TCGGAGGACGAGGTATGGG |
| Sequence-based reagent | Nedd4, R | This paper | PCR primer | GGTACGGATCAGCAGTGAACA |
| Sequence-based reagent | Nr1h3, F | This paper | PCR primer | CTCAATGCCTGATGTTTCTCCT |
| Sequence-based reagent | Nr1h3, R | This paper | PCR primer | TCCAACCCTATCCCTAAAGCAA |
| Sequence-based reagent | Nr1i3, F | This paper | PCR primer | ATATGGGCCGAGGAACTGTGT |
| Sequence-based reagent | Nr1i3, R | This paper | PCR primer | GGCGTGGAAATGATAGCCTGT |
| Sequence-based reagent | Nr2f6, F | This paper | PCR primer | GAGGACGATTCGGCGTCAC |
| Sequence-based reagent | Nr2f6, R | This paper | PCR primer | GTAATGCTTTCCACTGGACTTGT |
| Sequence-based reagent | Nr3c1, F | This paper | PCR primer | AGCTCCCCCTGGTAGAGAC |
| Sequence-based reagent | Nr3c1, R | This paper | PCR primer | GGTGAAGACGCAGAAACCTTG |
| Sequence-based reagent | Nr4a1, F | This paper | PCR primer | TTGAGTTCGGCAAGCCTACC |
| Sequence-based reagent | Nr4a1, R | This paper | PCR primer | GTGTACCCGTCCATGAAGGTG |
| Sequence-based reagent | Nr5a2, F | This paper | PCR primer | TGAGGAACAACTCCGGGAAAA |
| Sequence-based reagent | Nr5a2, R | This paper | PCR primer | CAGACACTTTATCGCCACACA |
| Sequence-based reagent | Nr6a1, F | This paper | PCR primer | CGCAACGGTTTCTGTCAGGAT |
| Sequence-based reagent | Nr6a1, R | This paper | PCR primer | GTTCAGCTCGATCATCTGGGA |
| Sequence-based reagent | Ppard, F | This paper | PCR primer | CTCATGAATGTGCCCCAGGT |
| Sequence-based reagent | Ppard, R | This paper | PCR primer | GTGCAGCAAGGTCTCACTCT |
| Sequence-based reagent | Rxrg, F | This paper | PCR primer | CATGAGCCCTTCAGTAGCCTT |
| Sequence-based reagent | Rxrg, R | This paper | PCR primer | CGGAGAGCCAAGAGCATTGAG |
| Sequence-based reagent | Rara, F | This paper | PCR primer | ATGTACGAGAGTGTGGAAGTCG |
| Sequence-based reagent | Rara, Reserve | This paper | PCR primer | ACAGGCCCGGTTCTGGTTA |
| Sequence-based reagent | Srebf1, F | This paper | PCR primer | GCAGCCACCATCTAGCCTG |
| Sequence-based reagent | Srebf1, R | This paper | PCR primer | CAGCAGTGAGTCTGCCTTGAT |
| Sequence-based reagent | MeCP2, F | This paper | ChIP-qPCR primer | TAAGTGACAGGAGTCACAGCG |
| Sequence-based reagent | MeCP2, R | This paper | ChIP-qPCR primer | TGGGACGTTGTATGTAACGGG |
| Sequence-based reagent | Nr1h3, F | This paper | ChIP-qPCR primer | CAGCACGTTGTAATGGAAGCC |
| Sequence-based reagent | Nr1h3, R | This paper | ChIP-qPCR primer | TAGCATTCAGTGGAGGGAAGG |
| Sequence-based reagent | Rara, F | This paper | ChIP-qPCR primer | CGATGAGTGGCAAGGTCTTT |
| Sequence-based reagent | Rara, R | This paper | ChIP-qPCR primer | ATAGCATAGCACCAGGGACAC |
| Sequence-based reagent | shRNA for Nr1h3 | This paper | shRNA sequence | CCTCAAGGACTTCAGTTACAA |
| Sequence-based reagent | shRNA for Rara | This paper | shRNA sequence | GAGCAGCAGTTCCGAAGAGAT |
| Other | B6. Cg-Speer6-ps1Tg (Alb-cre)21Mgn/J mice | The Jackson Laboratory | Cat# 003574 | Hepatocyte-specific Cre-transgenic mouse |
| Other | MeCP2flox/flox | Shanghai Model Organisms Center | Cat #NM-CKO-190001 | Mice carrying the targeted MeCP2 allele |
| Other | C57/BL6 | Guangdong Medical Laboratory Animal Center | Cat #17 | Wild-type mouse |
| Software, algorithm | GraphPad Prism | GraphPad Software | Version 8.0 | |
| Software, algorithm | ModFit | ModFit LT Software | Version 4.1 | https://www.vsh.com/products/mflt/index.asp |
| Software, algorithm | ImageJ | ImageJ software | https://imagej.nih.gov/ij/ |
Additional files
-
Supplementary file 1
For the GO enrichment analysis, the binding proteins of Mecp2 during the cell cycle reentry.
- https://cdn.elifesciences.org/articles/89912/elife-89912-supp1-v1.csv
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89912/elife-89912-mdarchecklist1-v1.pdf





