Coupling and uncoupling of midline morphogenesis and cell flow in amniote gastrulation
Figures

‘Polonaise movements’ persist under suppression of Wnt/PCP pathway.
(A) Testing model of relationships between the polonaise movements, primitive streak (PS) morphogenesis, and the Wnt/PCP pathway. A-P and L-R; anterior-posterior and left-right body axes, respectively. (B) Diagram of experimental set up. (C, D) In situ hybridizations for Brachyury (Bra) in control- and ΔDEP-GFP-misexpressing embryos. (E) Overlay of embryos hybridized for Bra (n=4 for each). (F) Cell number in PS [control 8.5±0.4 × 103 cells vs ΔDEP 8.8±1.2 × 103 cells, p=0.68, n=3 for each (two-tailed Student’s t test)]. (G-H’’’) Trace of cell flow path of electroplated epiblast cells with Flowtrace. The initiation of the polonaise movements was set as Time 0 (t=0). See also Video 1 and Figure 1—source data 1. (I-J’’’) Streamlines, visualizing averaged cell flows during each time period. (K-L’’’) Vorticity plots, displaying an averaged measure of the local rotation during each time period. Blue, clockwise; red, counter-clockwise rotation. Scale bars: 500μm.
-
Figure 1—source data 1
Vector field plots in control- and ΔDEP-GFP.
- https://cdn.elifesciences.org/articles/89948/elife-89948-fig1-data1-v1.tif
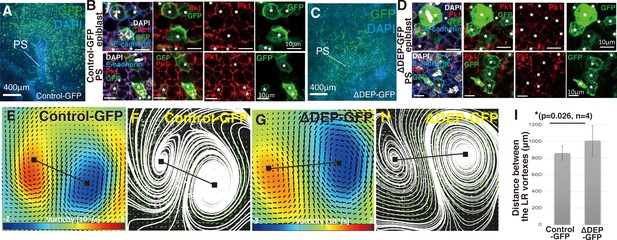
Distance between the LR rotations increases under suppression of the Wnt/PCP pathway.
(A, C) Immunofluorescent staining images of control- and ΔDEP-GFP-misexpressing PSs at HH3 with GFP (green) and DAPI (cyan). (B, D) Immunostaining images of epiblast and PS cells in control- and ΔDEP-GFP-misexpressing embryos at HH3 with GFP (green), Prickle1 (Pk1; magenta), and DAPI (blue). The white asterisks indicate GFP-expressing cells. (E, G) Averaged bilateral vortex-like-rotating cell flows (6 hr), visualized by vorticity plot, in control- and ΔDEP-GFP-misexpressing embryos. Black lines, measurement of the distance between the left-right (LR) rotations. See also Video 1 and Figure 1—source data 1. Blue, clockwise; red, counter-clockwise rotation. (F, H) Averaged bilateral vortex-like-rotating cell flows (6 hr), visualized by streamline plot, in control- and ΔDEP-GFP-misexpressing embryos. (I) Distance between the LR rotations in control- and ΔDEP-GFP-misexpressing embryos [control-GFP 860.2±86.2 μm vs ΔDEP-GFP 1008.4±183.8 μm, p=0.026, n=4 for each (two-tailed Student’s t test)].
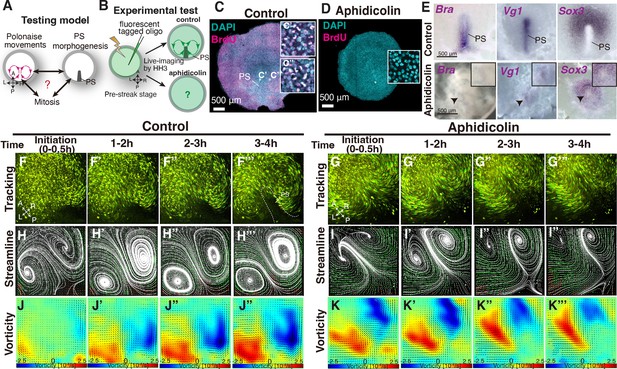
Early phase of ‘polonaise movements’ remains under global mitotic arrest.
(A) Testing model of relationships between the polonaise movements, primitive streak (PS) morphogenesis, and mitosis. Axes as in Figure 1. (B) Diagram of experimental set up. (C, D) BrdU incorporation in control-sham operated and aphidicolin treated in embryos. White boxes show enlarged areas. (C’, C”) High magnification of boxed areas in PS and non-PS, respectively. (E) In situ hybridizations for Brachyury (Bra), Vg1, and Sox3 in control and aphidicolin-treated embryos. Arrow heads indicate diminished PSs. Black boxes, enlarged area including the diminished PS. (F-G’’’) Trace of cell flow path with Flowtrace analysis (green is trace, yellow indicates endpoint) of fluorescently-tagged epiblast cells. The initiation of the polonaise movements was set as Time 0 (t=0). See also Video 2 and Figure 2—source data 1. (H-I’’’) Streamlines, visualizing averaged cell flows during each time period. Interpolated vectors are displayed in orange. (J-K’’’) Vorticity plots, displaying averaged measure of the local rotation during each time period. Blue, clockwise; red, counter-clockwise rotation. Scale bars: 500μm. Note: Since vorticity is calculated for all deviation from set point, slight curves and full rotations receive the same color indication, as seen in K’’’ which is not maintaining a bilateral vortex-like-rotating cell flows. See Materials and methods section for a full discussion.
-
Figure 2—source data 1
Vector field plots in control- and aphidicolin-treatment.
- https://cdn.elifesciences.org/articles/89948/elife-89948-fig2-data1-v1.tif
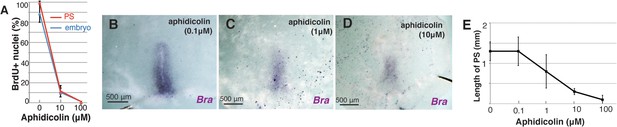
PS extension dose-dependently responds to aphidicolin.
(A) Dose-dependency of aphidicolin-induced mitotic arrest (PS, control 98.8±1.3% vs 10 uM aphidicolin 10.3±4.3% vs 100 uM aphidicolin 0 ± 0%, p=0.002e–4 in one-way ANOVA; embryo, control 87.9±9.0% vs 10 uM aphidicolin 9.0±4.4% vs 100 uM aphidicolin 0.02 ± 0.01%; n=4 for each; p=0.003e–5 in one-way ANOVA). (B–C) In situ hybridization for Brachyury (Bra) to different concentrations of aphidicolin treatment. (D) Length of PS at HH3 for different concentrations of aphidicolin treatment (control 1.3±0.2 mm vs 0.1 μM aphidicolin 1.3±0.3 mm vs 1 μM aphidicolin 0.8±0.4 mm vs 10 μM aphidicolin 0.3±0.1 mm vs 100 μM aphidicolin 0.1±0.1 mm; n=6 for each, p=6.2e–9 in one way ANOVA).
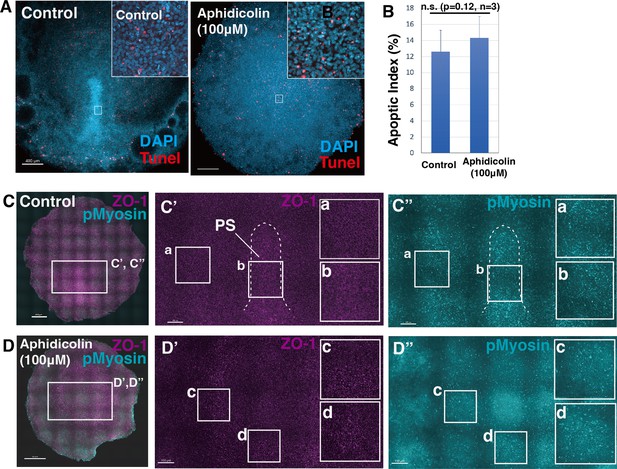
Aphidicolin-treatment maintains apoptotic index and pMyosin cables.
(A) TUNEL staining at stage HH 3 in control and aphidicolin-treated embryos. (B) Apoptotic index in control and aphidicolin-treated embryos (control 10.6±3.5% vs aphidicolin 12.1 ± 3.7%, p=0.20, n=4 for each). (C-D”) Immunofluorescent staining images of control- and aphidicolin-treated embryos at HH3 with ZO-1 (magenta) and pMyosin (cyan). White boxes show enlarged areas.
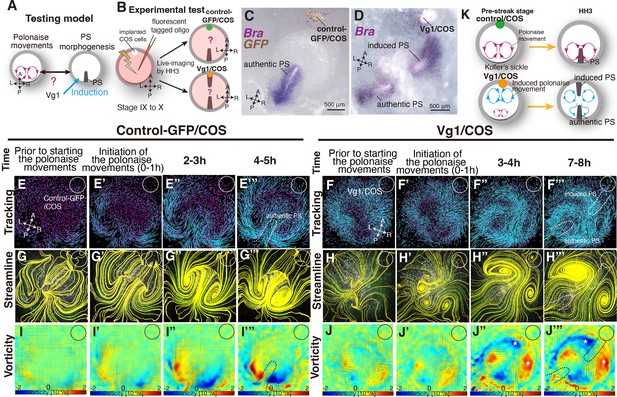
Authentic PS extends under disruption of authentic ‘polonaise movements’.
(A) Testing model of relationships between the polonaise movements, primitive streak (PS) morphogenesis, using ectopic Vg1-inducing system. Axes as in Figure 1. (B) Diagram of experimental set up. (C, D) In situ hybridizations for Brachyury (Bra) in control and ectopic Vg1-induced PS, respectively. (E-F’’’) Trace of cell flow path of electroplated epiblast cells by using Imaris tracking analysis. See also Video 3 and Figure 3—source data 1. White circles, COS cell implanted site. The initiation of the polonaise movements was set as Time 0 (t=0). (G-H’’’) Streamlines, visualizing averaged cell flows during each time period of the vector fields. (I-J’’’) Vorticity plots, displaying an averaged measure of the local rotation during each time period in the vector fields. Blue, clockwise; red, counter-clockwise rotation. Scale bars: 500μm. Note: vorticity is identified in both a full-rotation and curve, particularly in (J-J’’’). The white asterisks indicate the polonaise movements at the induced PS which are opposite direction to the authentic PS. (K) Summary of the cell flow patterns in control-GFP/COS- and Vg1/COS-implanted embryos, shown in (E-J’”).
-
Figure 3—source data 1
Vector field plots in control-GFP/COS and Vg1/COS.
- https://cdn.elifesciences.org/articles/89948/elife-89948-fig3-data1-v1.tif
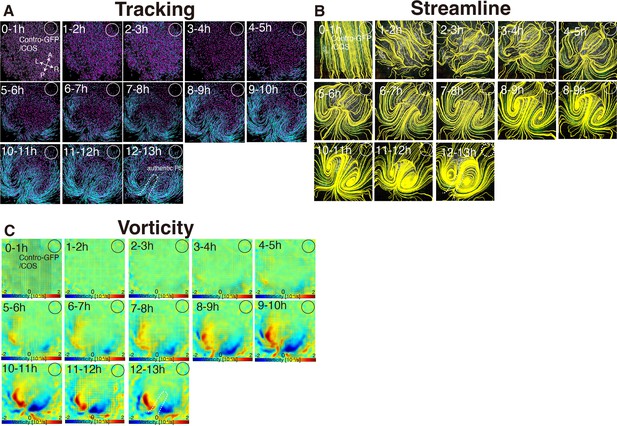
Time evolution of cell flow in control-GFP/COS-implanted embryo.
(A) Trace of cell flow path of electroplated epiblast cells by using Imaris tracking analysis. See also Video 3 and Figure 3—source data 1. A-P and L-R; anterior-posterior and left-right body axes, respectively. White circles, COS cell implanted site. (B) Streamlines, visualizing averaged cell flows during each time period. (C) Vorticity plots, displaying averaged measure of the local rotation during each time period. Blue, clockwise; red, counter-clockwise rotation.
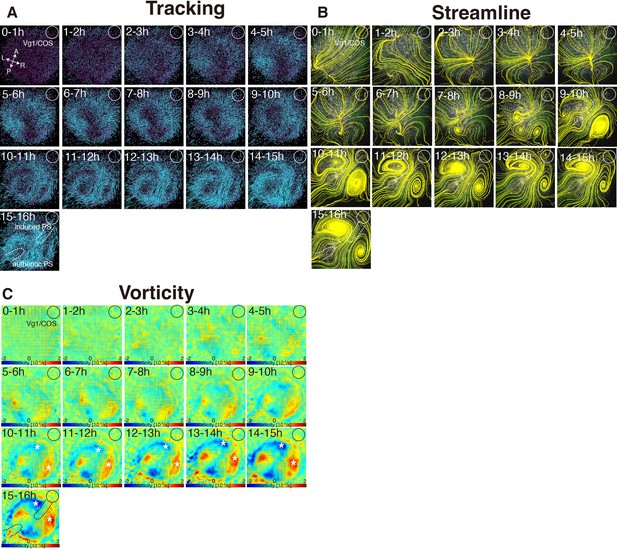
Time evolution of cell flow in Vg1/COS-implanted embryo.
(A) Trace of cell flow path of electroplated epiblast cells by using Imaris tracking analysis. See also Video 3 and Figure 3—source data 1. Axes are shown. White circles, COS cell implanted site. (B) Streamlines, visualizing averaged cell flows during each time period. (C) Vorticity plots, displaying averaged measure of the local rotation during each time period. Blue, clockwise; red, counter-clockwise rotation. White asterisks, the polonaise movements at the induced axis which are opposite direction to the authentic axis.

Induced ‘polonaise movements’ persist despite defective PS morphogenesis.
(A) Testing model of relationships between the polonaise movements, primitive streak (PS) morphogenesis, and Vg1. Axes as in Figure 1. (B) Diagram of experimental set up. (C, D) BrdU incorporation in control-GFP/COS- and Vg1/COS-implanted embryos under aphidicolin-treatment. White boxes showed enlarged areas. (E, F) In situ hybridizations for Brachyury (Bra, n=3 for each). Black arrows indicate diminished PSs. (G-H’’’) Trace of cell flow path of electroplated epiblast cells by using Imaris tracking analysis. See also Video 4 and Figure 4—source data 1. White circles, COS cell implanted site. The initiation of the polonaise movements was set as Time 0 (t=0). (I-J’’’) Streamlines, visualizing averaged cell flows during each time period of the vector fields. (K-L’’’) Vorticity plots, displaying an averaged measure of the local rotation during each time period. Note: vorticity is identified in both a full-rotation and curve, particularly in (L-L’’’). Blue, clockwise; red, counter-clockwise rotation. White asterisks, the polonaise movements at the induced axis which are opposite direction to the authentic midline axis. Scale bars: 500μm. Note: vorticity is identified in both a full-rotation and curve in (K’’’) and (L’’’). See description in the Materials and methods section.
-
Figure 4—source data 1
Vector field plots in control-GFP/COS and Vg1/COS under aphidicolin-treatment.
- https://cdn.elifesciences.org/articles/89948/elife-89948-fig4-data1-v1.tif
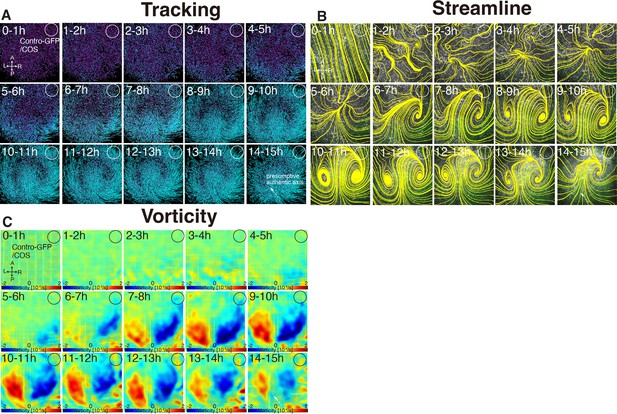
Time evolution of cell flow in control-GFP/COS-implanted embryo under aphidicolin-treatment.
(A) Trace of cell flow path of electroplated epiblast cells by using Imaris tracking analysis. See also Video 4 and Figure 4—source data 1. Axes are shown. White circles, COS cell implanted site. (B) Streamlines, visualizing averaged cell flows during each time period. (C) Vorticity plots, displaying averaged measure of the local rotation during each time period. Blue, clockwise; red, counter-clockwise rotation.
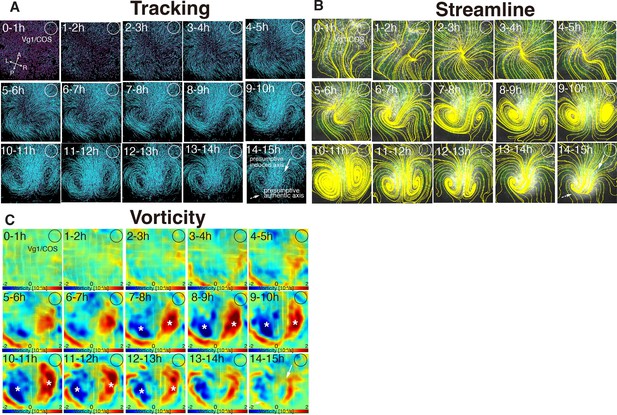
Time evolution of cell flow in Vg1/COS-implanted embryo under aphidicolin-treatment.
(A) Trace of cell flow path of electroplated epiblast cells by using Imaris tracking analysis. See also Video 4 and Figure 4—source data 1. Axes are shown. White circles, COS cell implanted site. (B) Streamlines, visualizing averaged cell flows during each time period. (C) Vorticity plots, displaying averaged measure of the local rotation during each time period. Blue, clockwise; red, counter-clockwise rotation. White asterisks, the polonaise movements at the induced axis which are opposite direction to the authentic axis.

Summary of the patterns of the cell flow and PS morphogenesis.
(A) Cell flow pattern under experimental manipulation of PS morphogenesis. (B) Authentic PS morphogenesis under disrupting authentic polonaise movements. (C) Vg1-induced cell flow pattern under defective PS morphogenesis. (D) Summary of the relationships between polonaise movements, PS morphogenesis, mitosis, and Wnt/PCP pathway.
Videos
‘Polonaise movements’ continue under suppression of Wnt/PCP pathway.
Time-lapse movie was analyzed by using Imaris tracking. Embryos were co-electroporated with lissamine-tagged oligo (red) and control-GFP or ΔDEP-GFP expression vectors (cyan) at pre-streak stage X-XII. Live imaging performed at 4 x objective lens on a spinning disk confocal microscope until HH3. Related to Figure 1, Figure 1—figure supplement 1, and Figure 1—source data 1. Scale bars, 300 μm.
Mitotic arrest maintains early phase of ‘polonaise movements’.
Time-lapse movie of fluorescently tagged epiblast cells in control and aphidicolin-treated embryos from pre-streak to HH3, processed by using Flowtrace. The trajectories, from green to yellow, indicate movement of fluorescently tagged cells (30 min projections). Embryos were electroporated with fluorescein-tagged oligo (green) and treated with DMSO (control-sham operated) or 100 μM of aphidicolin. Live-imaging performed at 4 x objective lends on a spinning disk confocal microscope for 10 hr. Related to Figure 2, Figure 2—figure supplement 2, Figure 2—figure supplement 2, and Figure 2—source data 1. Scale bars, 200 μm.
Authentic PS extends in absence of regular ‘polonaise movements’.
Embryos were electroporated with lissamine-tagged oligo (red) and implanted with COS cells expressing either a control-GFP (control-GFP/COS) or Vg1 (Vg1/COS) expression vector at anterior marginal zone at pre-streak stage IX-X. Live-imaging performed at a 2 x objective lens on a wide-field epifluorescent microscope. Magenta dots; lissamine-tagged epiblast cells, cyan lines; trajectories of the tagged epiblast cells for 2 hours. Related to Figure 3, Figure 3—figure supplement 1, Figure 3—figure supplement 2, and Figure 3—source data 1. Scale bars, 300 μm.
Ectopic Vg1 leads to ‘polonaise movements’ despite defective PS morphogenesis after mitotic arrest.
Embryos were electroporated with lissamine-tagged oligo (red), implanted with either control-GFP/COS or Vg1/COS at the anterior marginal zone (AMZ), and treated with 100 μM of aphidicolin at pre-streak stage IX to X. Live imaging performed at a 2 x objective lens with a wide-field epifluorescent microscope. Magenta dots; lissamine tagged epiblast cells, cyan lines; trajectories for 2.5 hr. Related to Figure 4, Figure 4—figure supplement 1, Figure 4—figure supplement 2, and Figure 4—source data 1. Scale bars, 300 μm.





