Serotonergic and dopaminergic neurons in the dorsal raphe are differentially altered in a mouse model for parkinsonism
Figures
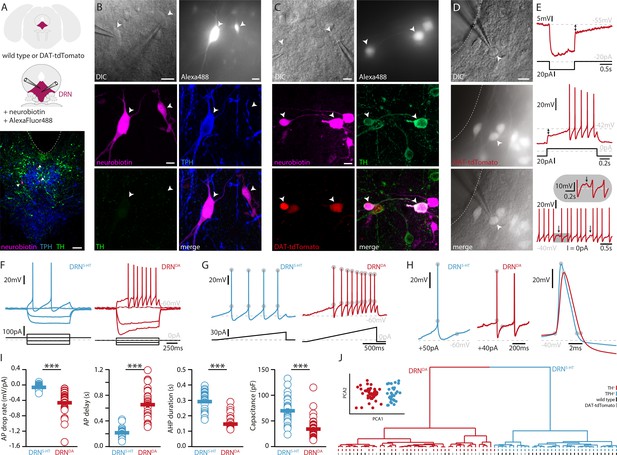
DRNDA and DRN5-HT are electrophysiologically distinct cell types.
(A) Scheme of the location of the DRN (pink) in a coronal section (top) and at higher magnification together with two patch pipettes (center). Bottom: a representative slice stained post-recording for TPH, TH, and neurobiotin revealing serotonergic neurons (arrows). The ventricle is indicated with a dashed line. (B) Top: differential interference contrast (DIC) microscopy image (left) of neurons that were filled with Alexa488 (right) and neurobiotin. Center, bottom: staining of the same neurons revealing a TPH+ (DRN5-HT) neuron and a TPH− and TH− cell. (C) Top: DIC image of recorded neurons that were filled with Alexa488 and neurobiotin. Center, bottom: staining of the same neurons revealing tdTomato+ and TH+ (DRNDA) neurons. (D) Representative fluorescent (top), DIC (center) image, and overlay (bottom) of a tdTomato+ neuron in a DAT-tdTomato mouse. (E) Representative recordings depicting postinhibitory hypoexcitability, slowly ramping currents and rebound oscillations in DRNDA neurons. (F) Representative voltage responses to current injections in a DRN5-HT and DRNDA neuron. (G) Ramping current injections reveal AP amplitude accommodation. Gray circles indicate the onset and peak of APs. (H) Amplitude and duration of the AP and AHP in a DRN5-HT and DRNDA neuron. Gray circles indicate onset, peak, and end of the AP and AHP. (I) Quantification of electrophysiological properties distinguishing DRN5-HT from DRNDA neurons (AP drop rate: n = 32 DRN5-HT, n = 43 DRNDA, Capacitance: n = 32 DRN5-HT, n = 43 DRNDA, AP delay: n = 30 DRN5-HT, n = 43 DRNDA, AHP duration: n = 28 DRN5-HT, n = 32 DRNDA, N = 9; Wilcoxon Rank Sum Test). (J) PCA of five electrophysiological parameters (insert) and hierarchical cluster analysis based on PCA1 and PCA2 (Ward’s method, Euclidean distance). Intrinsic properties were sufficient to separate TPH+ cells (blue dash) from TH+ (red dash) cells. Bottom dashes indicate WT (black) and DAT-tdTomato (gray) mice. Data are shown as mean ± SEM, ***p < 0.001. Scale bars: A, 100 μm; B–D, 10 μm.
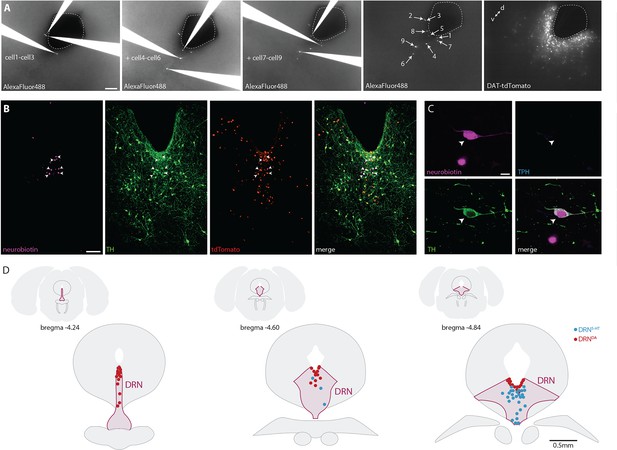
Filling of neurons with AlexaFluor488 and neurobiotin for subsequent immunohistochemistry and topographical registration.
(A) Snapshots of recorded neurons filled with AlexaFluor488 facilitate post hoc mapping of electrophysiology, anatomical location, and immunohistochemistry. Please note that cell 8 is shown at higher magnification in Figure 1D. (B) Representative confocal pictures of post-recording immunostaining for TH and neurobiotin revealing six TH+ neurons (arrows) in a DAT-tdTomato mouse. (C) Post-recording immunostaining of a TH+ neuron in a wild-type mouse. (D) Histological verification of recording position. Representative schemes showing the location of DRN5-HT and DRNDA neurons recorded in Sham. Neurons are plotted in the nearest coronal section (−4.24, −4.60, or −4.84 from Bregma). Scale bars: A, B, 100 µm; C, 10 µm; D, 0.5 mm.
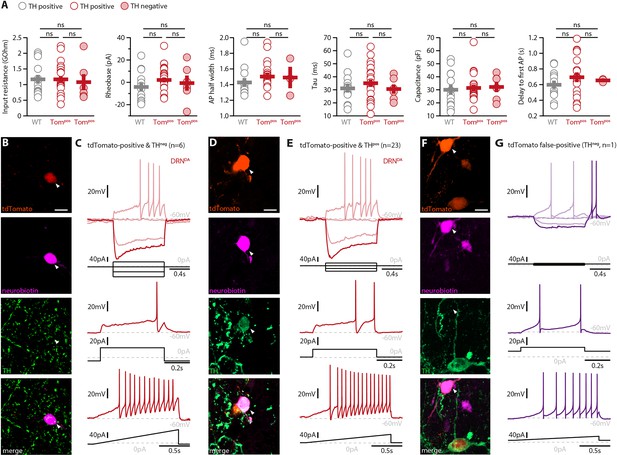
Electrophysiological properties of DRNDA neurons recorded in WT or DAT-tdTomato mice do not differ.
(A) Intrinsic properties of DRNDA recorded in WT or DAT-tdTomato (Tompos) mice (n = 13 DRNDA neurons from N = 5 WT mice vs. n = 29 DRNDA neurons from N = 3 tdTomato mice). Six of 29 tdTomato+ neurons were found to be TH− (closed circles) while all other neurons were TH+ (open circles). No statistically significant differences were found when comparing the electrophysiological properties of these three groups (ANOVA or Kruskal–Wallis test). (B) Post-recording immunohistochemistry showing the tdTomato-labeling, neurobiotin-filling, and absence of TH staining in a DRN neuron (indicated with a white arrow). (C) Whole-cell recording of the neuron shown in (B) and its responses to current injections. Hierarchical cluster analysis of the electrophysiological profile suggests that this neuron is a DRNDA neuron despite the absence of TH staining. (D) Post-recording immunohistochemistry showing a DRNDA neuron whose identity is confirmed by both TH staining and tdTomato-labeling (indicated with a white arrow). (E) Whole-cell recording of the neuron shown in (D) showing the classic electrophysiological profile of DRNDA neurons. (F) Post-recording immunohistochemistry showing the tdTomato-labeling, neurobiotin-filling, and absence of TH staining in a neuron false-positive for tdTomato (indicated with a white arrow). (G) Whole-cell recording of the neuron shown in (F) and its responses to different current injections. Please note the high input resistance, rebound spiking, biphasic afterhyperpolarization, and absence of spike amplitude accommodation. Scale bar: 10 µm.
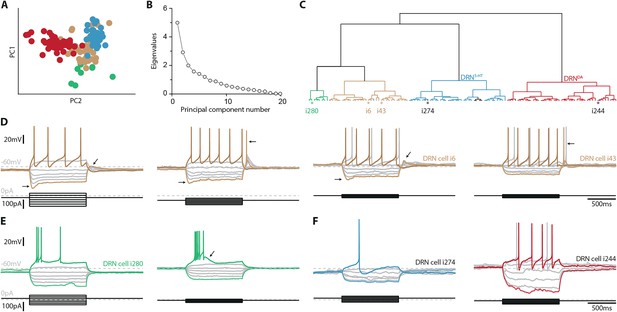
Clustering of DRN neurons based on electrophysiological parameters.
(A) PCA of 120 DRN neurons (N = 9) in Sham condition using 20 standard electrophysiological parameters. (B) Scree plot of the PCA. (C) Hierarchical cluster analysis (Ward’s method, Euclidean distance) based on the first three principal components. Colors indicate four main clusters. DRN5-HT neurons are indicated in blue, DRNDA neurons are indicated in red. Stars and corresponding identification numbers indicate DRN neurons of which example recordings are shown below. (D–F) Voltage responses to a series of current steps obtained from a variety of neurons in the DRN of Sham mice. (D) Four representative examples of the most frequently observed TH− and TPH− neurons recorded in the DRN. Please note the regular firing pattern, activation of sag currents (below −80 mV), and rebound depolarization/spiking. (E) Two examples of DRN neurons characterized by plateau potentials, bursting and spike frequency accommodation. (F) Examples of molecularly unidentified neurons that were assigned to the DRN5-HT (left) and DRNDA (right) cluster by the hierarchical cluster analysis.
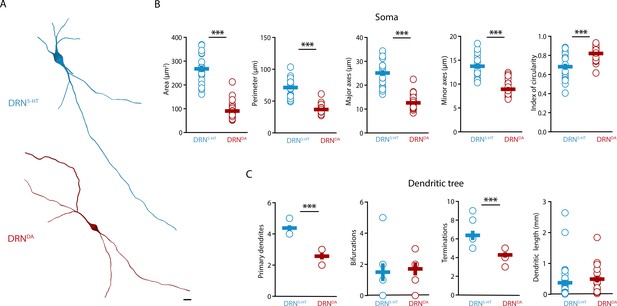
DRNDA and DRN5-HT have distinct morphological profiles.
(A) Top: representative digital reconstruction of a DRN5-HT. Bottom: representative digital reconstruction of a DRNDA. (B) Morphological parameters describing the soma size and shape of DRN5-HT and DRNDA neurons (DRN5-HT: n = 20, N = 3; DRNDA: n = 27, N = 3; unpaired t-test or Mann–Whitney U test). (C) Morphological parameters describing the dendritic tree of DRN5-HT and DRNDA neurons (DRN5-HT: n = 8, N = 3; DRNDA: n = 7, N = 3; Mann–Whitney U test). Data are shown as mean ± SEM, ***p < 0.001. Scale bar: 10 μm.
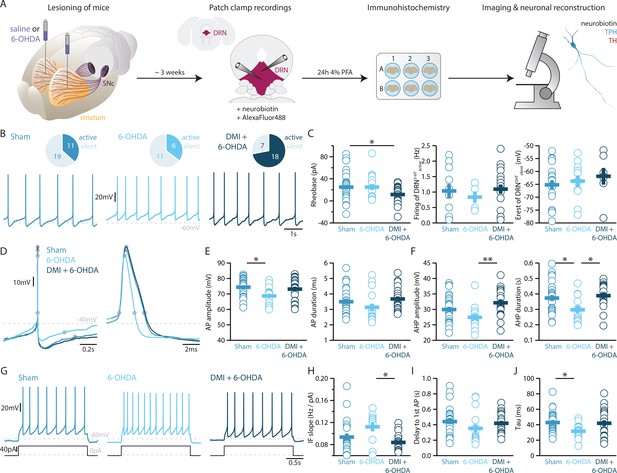
Lesions targeting primarily nigrostriatal dopamine increase the excitability of DRN5-HT neurons whereas loss of NA affects their APs.
(A) Overview of workflow for analyzing the electrophysiological and morphological properties of DRN neurons in Sham- and 6-OHDA-lesioned mice. (B) Top: pie charts showing the number of spontaneously active (dark) and silent (pale) DRN5-HT neurons in three conditions: Sham (left), 6-OHDA-injected mice (center), and 6-OHDA-injected mice pre-treated with DMI + 6-OHDA (right). Bottom: representative recordings of spontaneously active DRN5-HT neurons (I = 0 pA). (C) Quantification of the rheobase (left, Sham: n = 30, 6-OHDA: n = 17, DMI + 6-OHDA: n = 25), the firing frequency of spontaneously active cells (center, Sham: n = 11, 6-OHDA: n = 6, DMI + 6-OHDA: n = 18), and the resting membrane potential of silent DRN5-HT neurons (right, Sham: n = 19, 6-OHDA: n = 11, DMI + 6-OHDA: n = 7). (D) Representative APs of DRN5-HT at low (left) and high (right) temporal resolution. Gray circles indicate onset, offset, and peak of the APs as well as the end of the AHP. (E) Quantification of the amplitude (left) and duration (right) of the APs of DRN5-HT neurons (Sham: n = 29, 6-OHDA: n = 16, DMI + 6-OHDA: n = 21). (F) Same as in (D) for the AHP. (G) Representative responses of DRN5-HT neurons to current steps (I = +75 pA). (H) Quantification of firing frequency/injected current. (I) Quantification of the delay to the first AP when injected with current eliciting 1 Hz firing (Sham: n = 29, 6-OHDA: n = 16, DMI + 6-OHDA: n = 21). (J) Quantification of the membrane time constant tau of DRN5-HT neurons (Sham: n = 32, 6-OHDA: n = 16, DMI + 6-OHDA: n = 21). Sham: N = 6–7; 6-OHDA: N = 7; DMI + 6-OHDA: N = 4; unpaired t-test or Mann–Whitney U test. Data are shown as mean ± SEM, *p < 0.05, **p < 0.01.
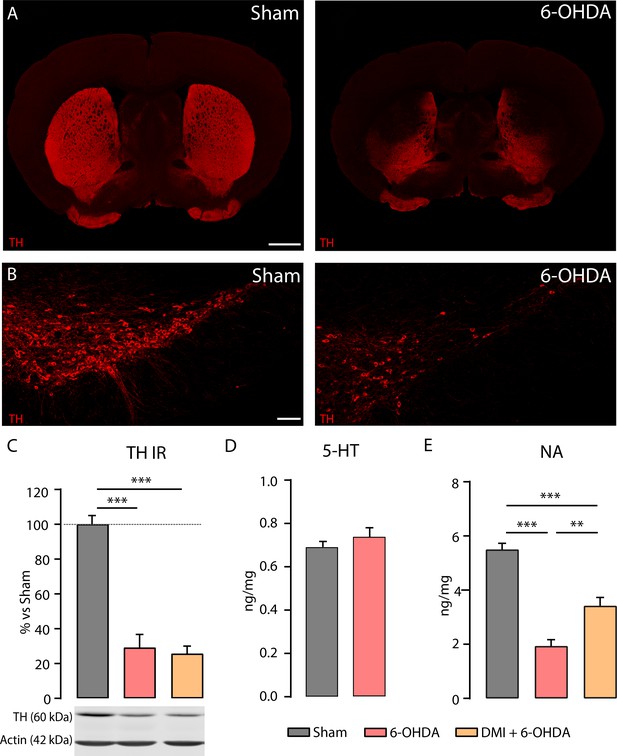
The striatal injection of 6-OHDA induced 60–70% TH loss in the striatum, did not alter striatal 5-HT levels but reduced striatal NA levels.
The reduction of striatal NA levels was partially prevented by the pre-treatment with DMI. (A) Representative confocal images in Sham (left) and 6-OHDA (right) showing the localization of TH loss in the dorsal striatum. (B) Representative confocal images in Sham (left) and 6-OHDA (right) showing the TH immunoreactivity in the SNc of Sham and 6-OHDA-injected mice. (C) Top: bar chart showing the TH levels in Sham, 6-OHDA, and DMI + 6-OHDA measured by western blot in the striatum (N = 6–9 per group: ANOVA). Data are normalized to Sham group (N = 6–9 per group; one-way ANOVA). Bottom: representative blots of TH and Actin immunoreactivity in Sham (left), 6-OHDA (center), and DMI + 6-OHDA (right). (D) Bar chart showing the 5-HT levels in Sham and 6-OHDA measured by ELISA in the striatum (N = 5–6 per group). (E) Bar chart showing the NA levels in Sham, 6-OHDA, and DMI + 6-OHDA measured by ELISA in the striatum (N = 9–10 per group; one-way ANOVA). Data are presented as mean ± SEM. ***p < 0.001, **p < 0.01. Scale bars: A, 1 mm; B, 100 µm.
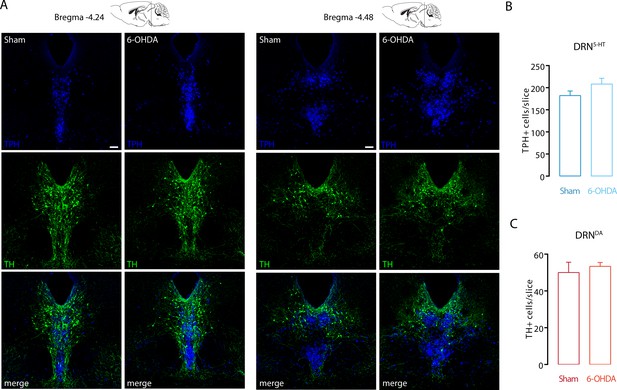
The 6-OHDA injection did not affect the number of DRN5-HT and DRNDA neurons.
(A) Representative confocal pictures of DRN5-HT and DRNDA in Sham and 6-OHDA mice at different antero-posteriorities. (B) Bar chart showing the density of TPH+ neurons in the DRN in Sham and 6-OHDA groups. (C) Bar chart showing the density of TH+ neurons in the DRN in Sham and 6-OHDA groups. N = 4 per group. Data are shown as mean ± SEM. Scale bar: 100 μm.
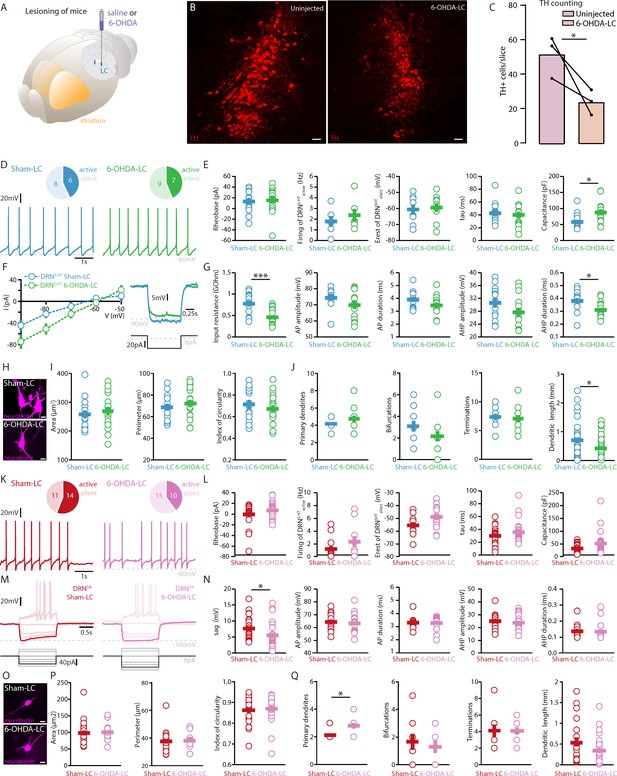
Selective lesioning of the NA system based on 6-OHDA injections in the LC affects DRN neurons mildly.
(A) Scheme showing unilateral injection of vehicle/6-OHDA in LC. (B) Representative confocal images of the lesion induced by the unilateral injection of 6-OHDA in the LC (left: uninjected side, contralateral to the 6-OHDA injection; right: 6-OHDA-injected side). (C) Bar chart showing the density of TH+ neurons in the LC in the uninjected and 6-OHDA-injected sides. Data are shown as mean ± SEM (N = 3 per group, unpaired t-test). (D) Top: pie charts showing the number of spontaneously active (dark) and silent (pale) DRN5-HT neurons in mice injected with vehicle (Sham-LC, left) and mice injected with 6-OHDA in LC (6-OHDA-LC, right). Bottom: representative recordings of spontaneously active DRN5-HT neurons (I = 0 pA). (E) Quantification of the rheobase (Sham-LC: n = 14, 6-OHDA-LC: n = 16), the firing frequency of spontaneously active cells (Sham-LC: n = 6, 6-OHDA-LC: n = 7), the resting membrane potential of silent DRN5-HT neurons (Sham-LC: n = 8, 6-OHDA-LC: n = 9), the time constant tau and the capacitance (Sham-LC: n = 14, 6-OHDA-LC: n = 15). (F) Representative responses of DRN5-HT neurons to hyper- (left) and depolarizing (right) current steps. (G) Quantification of the input resistance (Sham-LC: n = 14, 6-OHDA-LC: n = 16) and AP properties of DRN5-HT neurons (AP and AHP amplitude and duration: Sham-LC: n = 13, 6-OHDA-LC: n = 15). (H) Representative confocal pictures of soma from DRN5-HT neurons in Sham-LC (top) and 6-OHDA-LC mice (bottom). (I) Morphological descriptors of the soma size and shape in DRN5-HT neurons (Sham-LC: n = 15, N = 5, 6-OHDA-LC: n = 17, N = 5). (J) Morphological descriptors of the dendritic tree in DRN5-HT neurons. (Sham-LC: n = 11, N = 4; 6-OHDA-LC: n = 12, N = 4; Mann–Whitney U test). (K) Same as in (D) for DRNDA neurons. (L) Quantification of the rheobase (Sham-LC: n = 25, 6-OHDA-LC: n = 25), the firing frequency of spontaneously active cells (Sham-LC: n = 14, 6-OHDA-LC: n = 10), the resting membrane potential of silent DRNDA neurons (Sham-LC: n = 11, 6-OHDA-LC: n = 15), the time constant tau and the capacitance (Sham-LC: n = 25, 6-OHDA-LC: n = 25). (M) Representative responses of DRNDA neurons recorded in Sham-LC (left) and 6-OHDA-LC (right) to current steps. The step hyperpolarizing the neurons to −100 mV is highlighted. (N) Quantification of the sag amplitude (Sham-LC: n = 23, 6-OHDA-LC: n = 23) and AP properties of DRNDA neurons (AP and AHP amplitude and duration: Sham-LC: n = 15, 6-OHDA-LC: n = 18). (O) Representative confocal pictures of soma from DRNDA neurons in Sham-LC (top) and 6-OHDA-LC mice (bottom). (P) Morphological descriptors of the soma size and shape in DRNDA neurons (Sham-LC: n = 23, N = 5, 6-OHDA-LC: n = 23, N = 5). (Q) Morphological descriptors of the dendritic tree in DRNDA neurons (Sham-LC: n = 10, N = 3; 6-OHDA-LC: n = 10, N = 4; Mann–Whitney U test). All electrophysiological data were acquired in N = 5 Sham-LC and N = 5 6-OHDA-LC mice and statistics are based on unpaired t-test or Mann–Whitney U test. Data are shown as mean ± SEM, *p < 0.05,***p < 0.001. Scale bars: B, 50 μm, H–O, 10 μm.
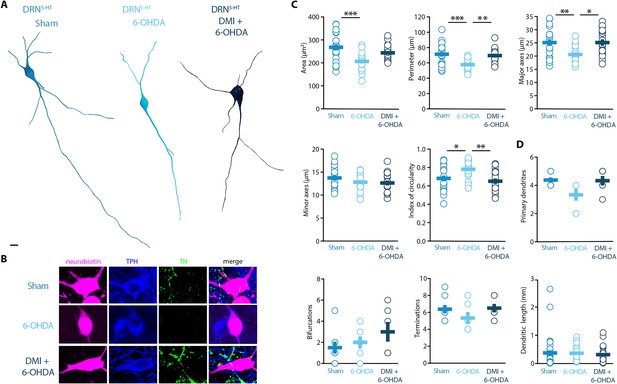
Striatal injection of 6-OHDA induced a hypotrophic phenotype in the DRN5-HT, which is prevented by pre-treatment with DMI.
(A) Representative digital reconstructions of a DRN5-HT neuron in three different conditions: Sham (left), 6-OHDA-injected mice (center), and 6-OHDA-injected mice pre-treated with DMI (right). (B) Representative confocal pictures of soma from DRN5-HT neurons in Sham (top), 6-OHDA-injected mice (center), and 6-OHDA-injected mice pre-treated with DMI (bottom). (C) Morphological descriptors of the soma size and shape in DRN5-HT neurons (Sham: n = 20, N = 4; 6-OHDA: n = 19, N = 4; DMI + 6-OHDA: n = 17, N = 3; one-way ANOVA). (D) Morphological descriptors of the dendritic tree in DRN5-HT neurons (Sham: n = 8, N = 3, 6-OHDA: n = 6, N = 3: DMI + 6-OHDA: n = 6, N = 2). Data are shown as mean ± SEM, ***p < 0.001, **p < 0.01, *p < 0.05. Scale bar: 10 µm.
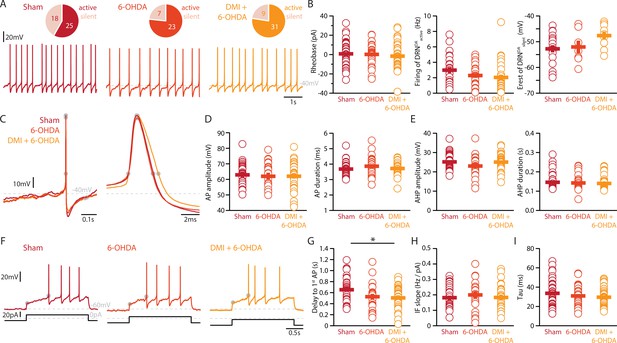
Lesions targeting primarily SN dopamine depolarize DRNDA neurons whereas concomitant loss of NA does not affect their APs.
(A) Top: pie charts showing the proportion of spontaneously active (dark) and silent (pale) DRNDA neurons in three conditions: Sham (left), 6-OHDA-injected mice (center), and 6-OHDA-injected mice pre-treated DMI (right). Bottom: representative recordings of spontaneously active DRNDA (I = 0 pA). (B) Quantification of the rheobase (left, Sham: n = 43, 6-OHDA: n = 31, DMI + 6-OHDA: n = 40), the firing frequency of spontaneously active (center, Sham: n = 25, 6-OHDA: n = 23, DMI + 6-OHDA: n = 31), and the resting membrane potential of silent DRNDA neurons (right, Sham: n = 18, 6-OHDA: n = 7, DMI + 6-OHDA: n = 9). (C) Representative APs of DRNDA at low (left) and high (right) temporal resolution. Gray circles indicate onset, offset, and peak of APs and the end of the afterhyperpolarization (AHP). (D) Quantification of the amplitude (left) and duration (right) of the APs of DRNDA neurons (Sham: n = 34, 6-OHDA: n = 23, DMI + 6-OHDA: n = 35). (E) Same as in (D) for the AHP. (F) Representative responses of DRNDA neurons to current steps (I = 75 pA). Gray circles indicate the delay to the first AP. Quantification of firing frequency/injected current (G, Sham: n = 31, 6-OHDA: n = 23, DMI + 6-OHDA: n = 27), the delay to the first AP when injected with current eliciting 2 Hz firing (H, Sham: n = 34, 6-OHDA: n = 23, DMI + 6-OHDA: n = 35), and the membrane time constant (I, Sham: n = 43, 6-OHDA: n = 29, DMI + 6-OHDA: n = 40) of DRNDA neurons recorded (Sham: N = 8; 6-OHDA: N = 6; DMI + 6-OHDA: N = 6; unpaired t-test or Mann–Whitney U test). Data are shown as mean ± SEM, *p < 0.05.
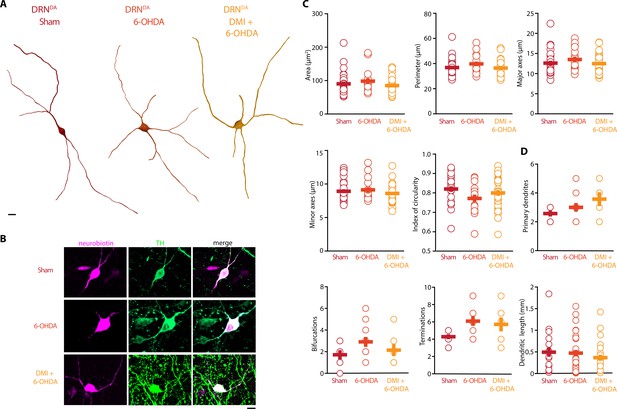
Striatal injection of 6-OHDA did not alter morphology of DRNDA.
(A) Representative digital reconstructions of a DRNDA neuron in three different conditions: Sham (left), 6-OHDA-injected mice (center), and 6-OHDA-injected mice pre-treated with DMI (right). (B) Representative confocal pictures of soma from DRNDA neurons in Sham (top), 6-OHDA-injected mice (center), and 6-OHDA-injected mice pre-treated with DMI (bottom). (C) Morphological descriptors of the soma size and shape in DRNDA neurons (Sham: n = 27, N = 7; 6-OHDA: n = 16, N = 4; DMI + 6-OHDA: n = 31, N = 5). (D) Morphological descriptors of the dendritic tree in DRNDA neurons (Sham: n = 7, N = 3; 6-OHDA: n = 11, N = 4; DMI + 6-OHDA: n = 7, N = 3). Data are shown as mean ± SEM. Scale bar: 10 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (mouse, C57BL/6J) | DAT-cre | The Jackson Laboratory | Stock #006660 | |
| Strain, strain background (mouse, C57BL/6J) | tdTomato | The Jackson Laboratory | Stock #007909 | |
| Antibody | anti-Tyrosine Hydroxylase (rabbit polyclonal) | Millipore | Millipore: AB152; RRID:AB_390204 | 1:1000 IF; 1:2000 WB |
| Antibody | anti-Tryptophane Hydroxylase (mouse monoclonal) | Sigma-Aldrich | Sigma-Aldrich: T0678; RRID:AB_261587 | 1:600 |
| Antibody | anti-Beta-Actin (mouse monoclonal) | Sigma-Aldrich | Sigma-Aldrich: A5316; RRID:AB_476743 | 1:30,000 |
| Commercial assay or kit | Noradrenaline Research ELISA kit | LDN | BA E-5200R | |
| Commercial assay or kit | Serotonine Research ELISA kit | LDN | BA E-5900R | |
| Chemical compound, drug | Desipramine hydrochloride | Sigma-Aldrich | D3900 | |
| Chemical compound, drug | 6-Hydroxydopamine hydrocloride | Sigma-Aldrich | H4381 | |
| Chemical compound, drug | Sucrose | Fisher Scientific | 10638403 | |
| Chemical compound, drug | Glucose | Sigma-Aldrich | G7021 | |
| Chemical compound, drug | NaHCO3 | Fisher Scientific | 10118190 | |
| Chemical compound, drug | KCl | Sigma-Aldrich | P3911 | |
| Chemical compound, drug | NaH2PO4 | Sigma-Aldrich | 71504 | |
| Chemical compound, drug | CaCl2 | Sigma-Aldrich | C5080 | |
| Chemical compound, drug | MgCl2 | Sigma-Aldrich | M2670 | |
| Chemical compound, drug | NaCl | Merck | 106404 | |
| Chemical compound, drug | K-gluconate | Sigma-Aldrich | G4500 | |
| Chemical compound, drug | HEPES | Sigma-Aldrich | H3375 | |
| Chemical compound, drug | Mg-ATP | Sigma-Aldrich | A9187 | |
| Chemical compound, drug | GTP | Sigma-Aldrich | G8877 | |
| Chemical compound, drug | Na2-phosphocreatine | Sigma-Aldrich | P7936 | |
| Chemical compound, drug | Neurobiotin | Vector Laboratories, Bionordika | SP-1120 | |
| Chemical compound, drug | AlexaFluor488 Hydrazide | Invitrogen/Thermo Fisher Scientific | A10436 | |
| Software, algorithm | Igor Pro 6.37 | Wavemetrics | RRID:SCR_000325 | |
| Software, algorithm | GraphPad Prism | Graphpad Software | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | Java | RRID:SCR_003070 | |
| Software, algorithm | neuTube | Howard Hughes Medical Institute; Feng et al., 2015 | RRID:SCR_024867 | |
| Other | Cy5-conjugated streptavidin | Jackson ImmunoResearch | Jackson ImmunoResearch: 016-170-084; RRID:AB2337245 | 1:500 |
| Other | NEUROBIOTIN Tracer | Vector Laboratories | Vector Laboratories: SP-1120; RRID:AB2313575 |





