Fungal–bacteria interactions provide shelter for bacteria in Caesarean section scar diverticulum
Figures

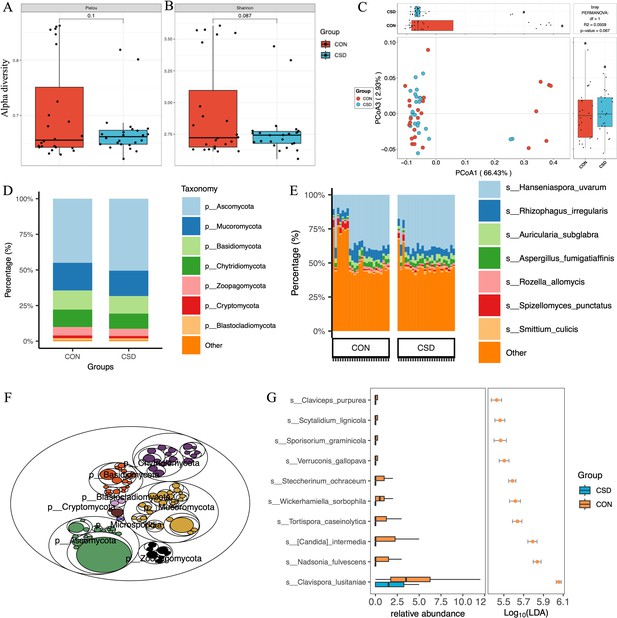
Bacterial community structure and composition characteristics.
Alpha diversity based on Pielou index (A) and Shannon index (B); (C) beta diversity based on the Bray–Curtis distance; stacked graph of community state type (CST) (D) and subCST (E) compositions; (F) stacked plot of species composition; (G) scatter plot of differentially abundant species between CSD and CON groups.
Fungal community structure and composition characteristics.
Alpha diversity based on Pielou index (A) and Shannon index (B); (C) beta diversity based on the Bray–Curtis distance; stacked graph of phylum level (D) and species level (E) compositions; (F) the tree map of fungal community; (G) box plot of differentially abundant species between CSD and CON groups.
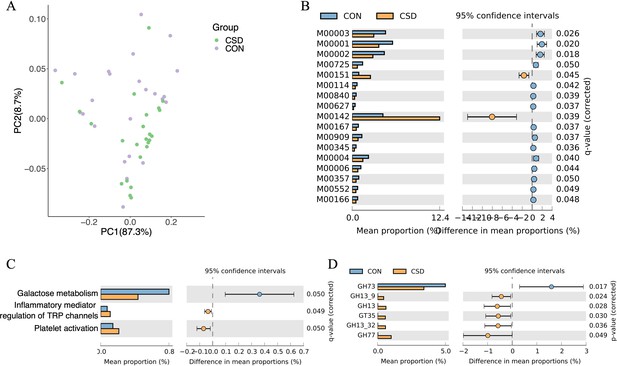
Functional composition of the cervical microbiota.
(A) PCA plot of microbial functional genes; (B) differences in the composition of Kyoto Encyclopedia of Genes and Genomes (KEGG) modules between CSD and CON; (C) differences in the composition of KEGG pathway between CSD and CON; (D) differences in the composition of carbohydrate-active enzymes between CSD and CON.
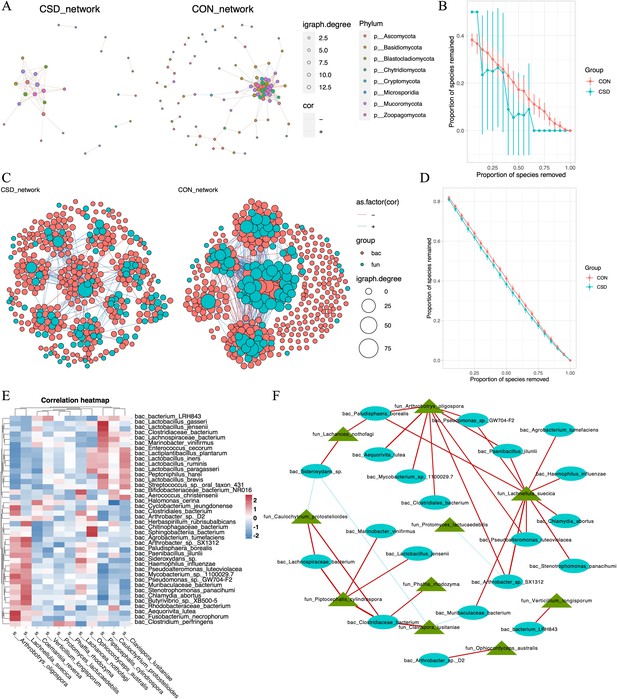
Interaction between bacteria and fungi.
(A) Co-occurrence network of fungal communities; (B) comparison of the random robustness of co-occurrence network of fungal communities between CSD and CON groups; (C) interaction network between bacteria and fungi; (D) comparison of the random robustness of bacterial–fungal interaction network between CSD and CON groups; (E) heatmap of the correlation between bacteria and fungi; (F) network of correlations between bacteria and fungi (R ≥ 0.6, p ≤ 0.05).
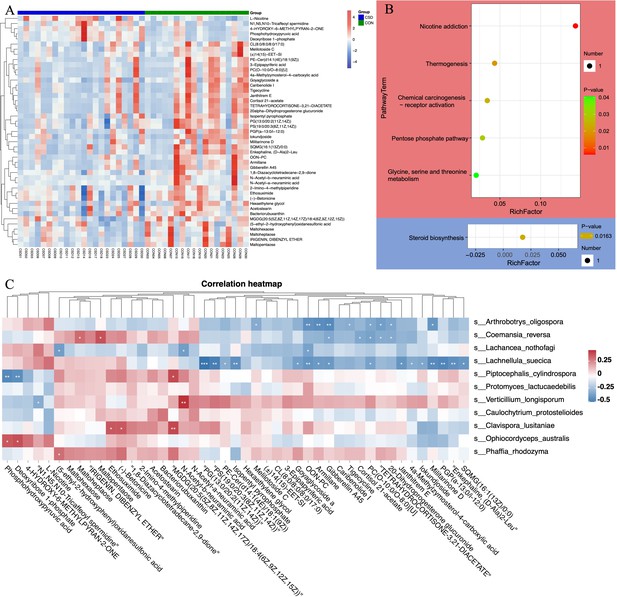
Metabolite composition and functional characteristics.
(A) Heatmap of differentially abundant metabolites between CSD and CON groups. (B) Dot plot of KEGG enrichment analysis of metabolites. Red represents upregulation, blue represents downregulation. (C) Heatmap of the correlation between fungi and metaboliteds.
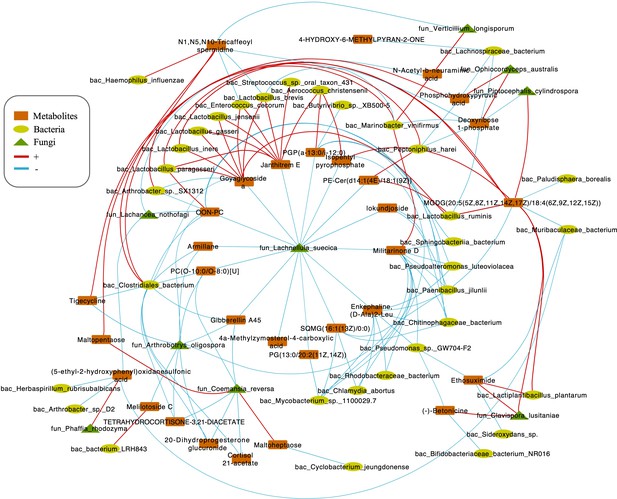
Correlations network of fungi, bacteria, and metabolites.
The orange module represents metabolites. The yellow module represents bacteria. The green module represents fungi. The red line represents positive correlations. Blue line represents negative correlations. The thickness of the lines represents the magnitude of the correlation coefficient.
Tables
Clinical characteristics of the participants included in the study.
| CON group (n = 24) | CSD group (n = 24) | p value | |
|---|---|---|---|
| AMH (ng/ml) (mean (SD)) | 3.69 (2.09) | 3.00 (2.32) | 0.288 |
| BMI (mean (SD)) | 21.04 (2.25) | 22.25 (2.70) | 0.1 |
| Basal FSH (IU/l) (mean (SD)) | 6.99 (1.42) | 7.70 (1.88) | 0.144 |
| Basal LH (IU/l) (mean (SD)) | 5.51 (2.97) | 5.91 (3.55) | 0.676 |
| Basal E2 (pg/ml) (mean (SD)) | 47.42 (68.52) | 44.49 (29.46) | 0.848 |
| infertile years (mean (SD)) | 3.54 (2.19) | 4.54 (2.90) | 0.184 |
Additional files
-
Supplementary file 1
431 fungal species in all samples.
- https://cdn.elifesciences.org/articles/90363/elife-90363-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/90363/elife-90363-mdarchecklist1-v1.docx