New genetic tools for mushroom body output neurons in Drosophila
Figures
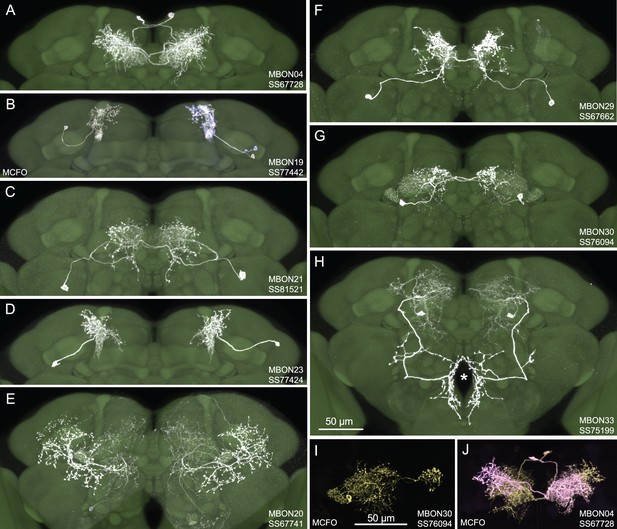
Selected images of new split-GAL4 lines.
Panels A and C–H show expression (maximum intensity projections) of the indicated split-GAL4 line in the relevant portion of the brain. In panels A–H, the consensus JFRC2018 unisex brain template is also shown (green). Images showing the full brain, optic lobe, and ventral nerve cord of these lines can be found in Figure 1—figure supplement 1 (for E–H) and Figure 1—figure supplement 2 (for A–D). Panels B, I, and J show images derived from stochastic labeling that reveal the morphology of individual cells. The original confocal stacks from which these images were derived are available for download at https://splitgal4.janelia.org/cgi-bin/splitgal4.cgi.
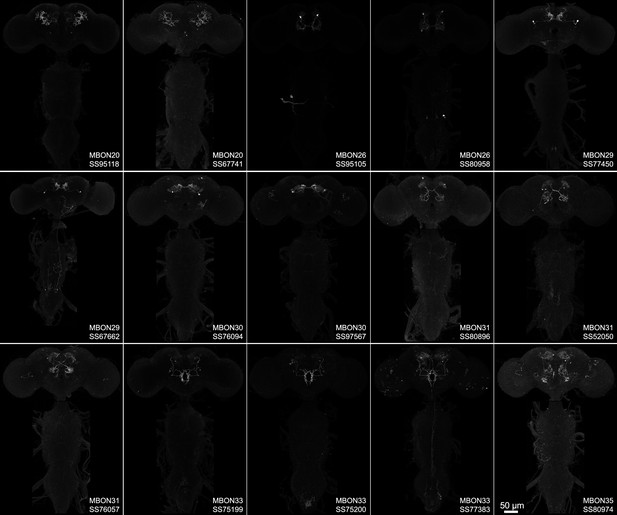
Maximum intensity projections of the brains and ventral nerve cords of split-GAL4 lines for atypical mushroom body output neuron (MBON).
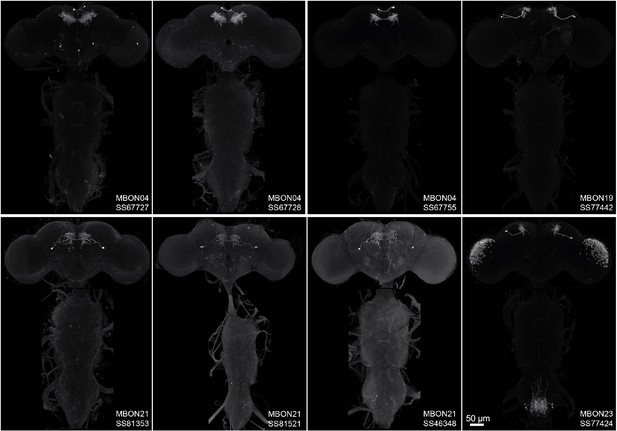
Maximum intensity projections of the brains and ventral nerve cords of split-GAL4 lines for typical mushroom body output neuron (MBON).
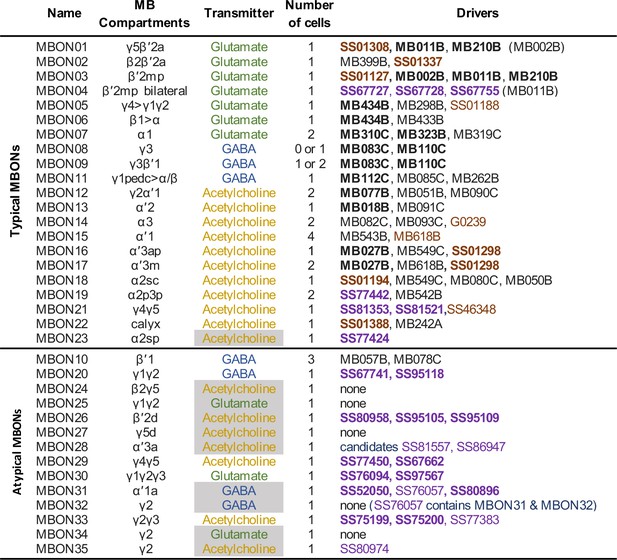
Summary list of selected split-GAL4 lines for all mushroom body output neuron (MBON) cell types.
These lines represent what we believe to be the best available split-GAL4 drivers for each cell type. The mushroom body (MB) compartments they innervate and neurotransmitters are shown. The neurotransmitters that are shaded in gray were assigned based solely on computational prediction (Eckstein et al., 2020). Other transmitters have been confirmed by antibody staining (Aso et al., 2014a) or EASI-FISH (Eddison and Ihrke, 2022). The number of cells per hemisphere is shown for each cell type. Available split-GAL4 are listed. Lines listed in boldface are generally of higher quality. Lines whose names are shown in purple font were generated as part of this study. Lines whose names are shown in black font were described in Aso et al., 2014a. Lines from other studies are shown brown font and their sources were as follows: SS01308, Aso et al., 2019; G0239, Chiang et al., 2011; SS46348, Otto et al., 2020; MB618B, SS01127, SS01188, SS01194, SS01298, SS01337, and SS01388, Shuai et al., 2023 release: https://splitgal4.janelia.org.
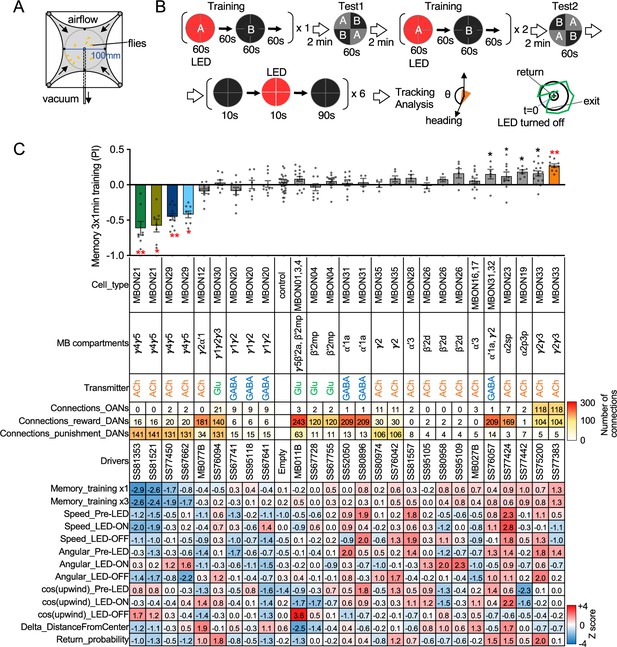
Behavioral consequences of optogenetic activation.
(A) The four-armed olfactory arena. Approximately 20 starved female flies were confined in 10 cm diameter and 3 mm high circular area with a hole at the center for air suction. Odor was introduced through four channels at the corners. (B) The protocol for behavioral experiments. Flies were trained by pairing 60 s of odor A with 30 1 s pulses of 627 nm LED light, each separated by 1 s without illumination. A different odor, odor B, was presented without red LED illumination, and then preference between two odors was tested. In the reciprocal experiments, odor B was paired with red light and A was unpaired. The same training was repeated twice more and then a second odor preference test was performed. Finally, six cycles of 10 s 627 nm illumination were applied, spaced by 100 s intervals without odor. Airflow was maintained at 400 mL/min throughout the experiment. (C) Top: The memory scores at the second odor preference test, measured as preference indexes: [(number of flies in the paired odor quadrants)-(number of flies in the unpaired odor quadrants)]/total number of flies during the last 30 s of the 60 s test period. The red asterisks * and ** indicate p<0.05 or p<0.01, respectively: Dunn’s multiple comparison tests compared to empty-split-GAL4 control, following Kruskal-Wallis test. The black * indicates p<0.05 without correction for multiple comparison. N=34 for the empty-split-GAL4 line and N=4–16 for other lines. All the lines were initially tested for four reciprocal experiments; lines with mean preference index above 0.1 or below –0.1 were subjected to additional tests. Cell types, the mushroom body (MB) compartments in which their dendrites lie, their neurotransmitters, the number of synaptic connections they make with dopaminergic (DANs) and octopaminergic (OANs) neurons, and the split-GAL4 driver lines used for the behavioral assays are designated. A summary of connections from all mushroom body output neuron (MBON) subtypes to DANs thought to signal reward or punishment and to OANs is shown in Figure 2—figure supplement 1A. Bottom: Z-scores [(values-mean)/standard deviation] for each parameter: speed, walking speed; angular, absolute of angular change relative to the previous frame at 30 FPS; cos(upwind), cosine of the fly’s orientation toward the upwind direction (i.e. facing away from the center of the arena). ON periods correspond to the first 2 s of the 10 s LED ON periods, whereas OFF periods are the 2 s immediately after the LEDs were turned off. Delta_DistanceFromCenter is change in fly’s mean distance from the center of the arena relative to its position at the onset of LED illumination. Return is a measure of the probability that flies return to the position that they occupied at the end of the LED stimulus. Flies are judged to have returned if they move 10 mm or more from their original position and then return to within 3 mm of the original position within 15 s.
-
Figure 2—source data 1
The values used for Figure 2.
- https://cdn.elifesciences.org/articles/90523/elife-90523-fig2-data1-v1.xlsx
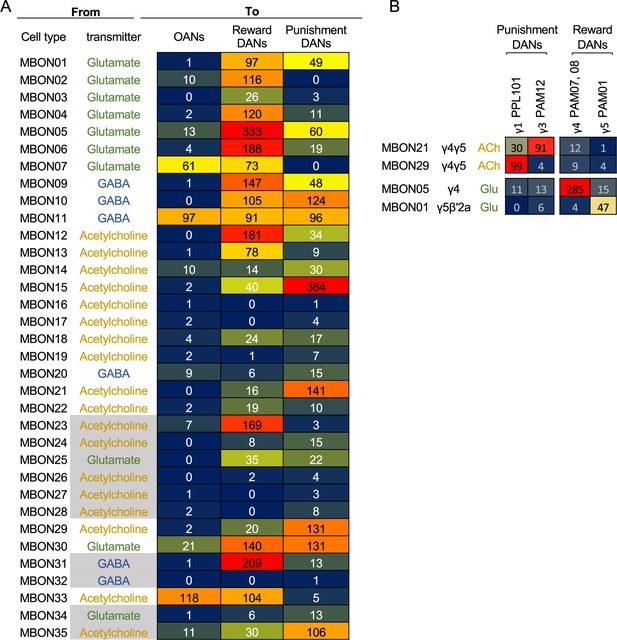
Direct connections from mushroom body output neurons (MBONs) to dopaminergic neurons (DANs) and octopaminergic neurons (OANs).
(A) Total number of synaptic connections from each MBON type to DANs and OANs. Based on the valence of memory when activation of DANs is used as unconditioned stimulus in olfactory conditioning (Aso et al., 2012; Aso et al., 2010; Aso and Rubin, 2016; Claridge-Chang et al., 2009; Huetteroth et al., 2015; Ichinose et al., 2015; Lin et al., 2014; Liu et al., 2012; Yamada et al., 2023; Yamagata et al., 2016Yamagata et al., 2015), we considered PAM01, 02, 04, 05, 06, 07, 08, 09, 10, 11, and 15 as Reward DANs and PAM12, 13, 14, PPL101, 103, and 106 as Punishment DANs. OANs refer to the four types of octopaminergic neurons that innervate the MB: OA-VPM3, OA-VPM4, OA-VUMa2, and OA-VUMa7. (B) Number of connections from MBONs in γ4 and γ5 to DANs in the γ lobe. PPL103, a DAN in the γ2 compartment, has less than five connections with these MBONs and was not included in this table.
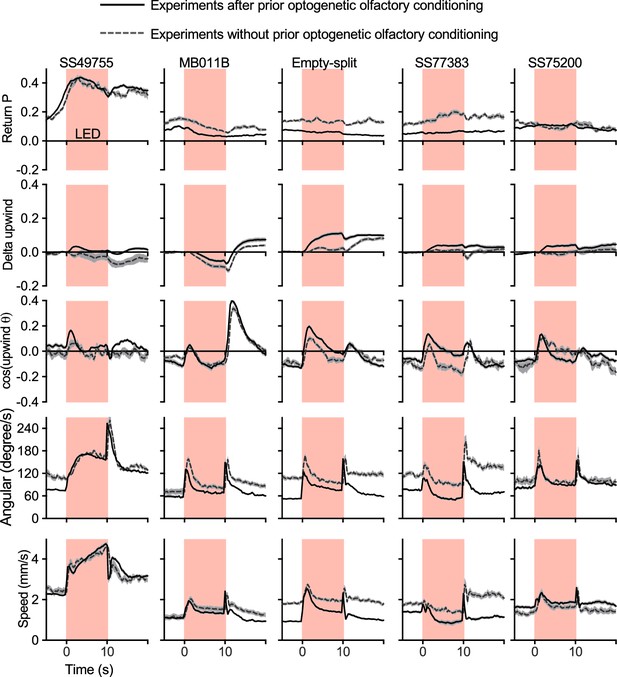
Activation phenotypes are not compromised by prior optogenetic olfactory conditioning.
Time course of five kinematic parameters shown in Figure 2 for the positive control lines (MB011B and SS49755), negative control (Empty-split-GAL4), and two lines for MBON33 (SS75200 and SS77383) are shown. Lines and dashed lines represent mean of six trial averages for experiments with or without prior optogenetic conditioning sessions as shown in Figure 2B. Lines and shadings are means and SEMs.
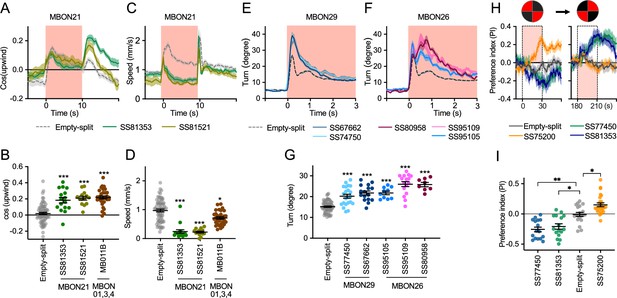
Additional behavioral consequences of optogenetic activation.
(A) Time course of mean cos(upwind angle) for flies that express CsChrimson in MBON21 with designated drivers. The trace of empty-split-GAL4 is also shown. All the trajectories from six trials of movies were pooled to calculate a mean for each group of flies. Lines and shadings are means and SEMs. (B) Mean cos(upwind angle) during 2 s periods immediately after LED was turned off. (C–D) Time course and mean walking speed during 10 s LED ON period. (E–F) The mean cumulative turning angles in five movie frames of (total elapsed time 150 ms) for flies expressing CsChrimson in MBON29 and MBON26. (G) The cumulative turning angle during the first 2 s of LED ON period. (A–G) show data from the experiments described in Figure 2. (H) Preference for quadrants with red light. Flies expressing CsChrimson in MBON21, MBON29, or MBON33 were tested with 30 s continuous light of 627 nm LED in two quadrants. The test was performed a second time with illumination in opposite quadrants after a 150 s recovery period. (I) Mean preference index to the quadrants with red light during the last 5 s of two 30 s test periods. Dunn’s multiple comparison tests compared to empty-split-GAL4 control, following Kruskal-Wallis test. *, **, and *** indicate p<0.05, p<0.01, or p<0.001, respectively: N=66 for Empty-split-GAL4 and 8–22 for other lines in (A–G). N=16–26 in (H–I).
-
Figure 3—source data 1
The values used for Figure 3.
- https://cdn.elifesciences.org/articles/90523/elife-90523-fig3-data1-v1.xlsx
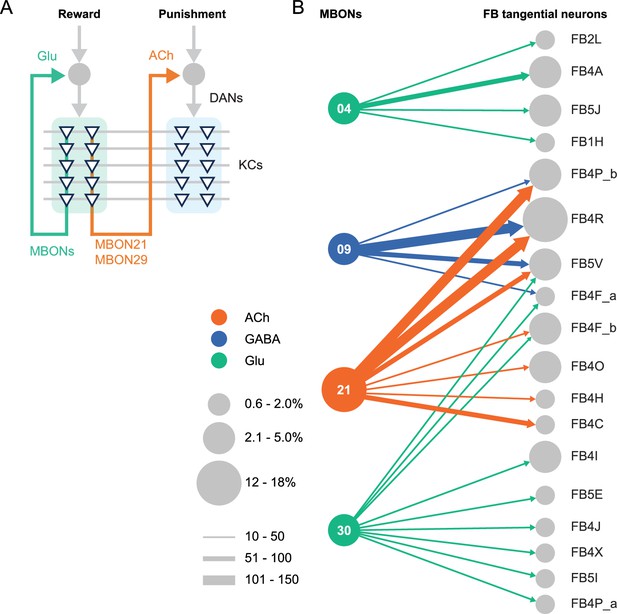
Diagrammatic summary of key outputs from selected mushroom body output neurons (MBONs).
(A) MBON21 and MBON29 arborize dendrites in the g4 and g5 compartments that are innervated by reward representing dopaminergic neurons (DANs). MBON21 and MBON29 are cholinergic and preferentially connect with DANs that innervate other compartments to represent punishment, whereas other glutamatergic MBONs from these same compartments preferentially form connections with reward-representing DANs going back to the same compartments. In fly brains, acetylcholine (ACh) is largely excitatory via nicotinic ACh receptors, although the type A muscarinic ACh receptor can mediate an inhibitory effect (Bielopolski et al., 2019; Manoim et al., 2022). Glutamate (Glu) can be inhibitory or excitatory depending on the receptors in the postsynaptic cells. Glutamate is known to be inhibitory via the glutamate-gated chloride channel (GluClα) in the olfactory system (Liu and Wilson, 2013). All of the 10 types of DANs examined with RNA-seq express GluClα and Nmdar2 at high levels whereas expression of Nmdar1 and other glutamate receptors were limited and cell type specific (Aso et al., 2019). Results in some studies support an excitatory effect of at least a subset of glutamatergic MBONs on DANs (Cohn et al., 2015; Ichinose et al., 2015; Otto et al., 2020; Zhao et al., 2018a), while electrophysiological recordings identified inhibitory connection between glutamatergic MBON and the downstream interneurons (Aso et al., 2023). (B) Diagram showing direct connections between the mushroom body (MB) and central complex (CX) mediated by MBON21, MBON30, MBON09, and MBON04; these MBONs rank first, second, fifth, and seventh, respectively, based on the number of direct synaptic connections to the CX; numbers reflect connections between right hemisphere MBONs and right hemisphere FB tangential cells. For circles representing MBONs, the circle diameter represents the fraction of that MBONs direct output that goes to the CX. For the downstream neurons in the CX, circle diameter represents the fraction of that cell types direct input that comes from MBONs. Arrow width reflects synapse number. See Figure 19 of Li et al., 2020 and Figure 46 of Hulse et al., 2021 for additional information on the complete set of MB to CX connections.
Videos
Comparison of light microscopic images of atypical mushroom body output neurons (MBONs) with hemibrain skeletons of the corresponding cell types.
Comparison of light microscopic images of typical mushroom body output neurons (MBONs) with hemibrain skeletons of the corresponding cell types.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Drosophila melanogaster) | Split-GAL4 lines | This paper, Aso et al., 2014a; PMID: 25535793 | https://splitgal4.janelia.org/cgi-bin/splitgal4.cgi | Available from Aso lab and Rubin lab |
| Strain, strain background (Drosophila melanogaster) | 20xUAS-CsChrimson- mVenus attP18 | Klapoetke et al., 2014; PMID: 24509633 | N.A. | |
| Strain, strain background (Drosophila melanogaster) | pJFRC200-10xUAS- IVS-myr::smGFP-HA in attP18 | Nern et al., 2015; PMID: 25964354 | N.A. | |
| Strain, strain background (Drosophila melanogaster) | pJFRC225-5xUAS- IVS-myr::smGFP-FLAG in VK00005 | Nern et al., 2015; PMID: 25964354 | N.A. | |
| Strain, strain background (Drosophila melanogaster) | pBPhsFlp2::PEST in attP3 | Nern et al., 2015; PMID: 25964354 | N.A. | |
| Strain, strain background (Drosophila melanogaster) | pJFRC201-10XUAS-FRT>STOP > FRT-myr::smGFP-HA in VK0005 | Nern et al., 2015; PMID: 25964354 | N.A. | |
| Strain, strain background (Drosophila melanogaster) | pJFRC240-10XUAS-FRT>STOP > FRT-myr::smGFP-V5-THS-10XUAS-FRT>STOP > FRT-myr::smGFP-FLAG_in_su(Hw)attP1 | Nern et al., 2015; PMID: 25964354 | N.A. | |
| Strain, strain background (Drosophila melanogaster) | empty-split-GAL4 (p65ADZp attP40, ZpGAL4DBD attP2) | Hampel et al., 2015; PMID: 26344548 | RRID:BDSC_79603 | |
| Antibody | Anti-GFP (rabbit polyclonal) | Invitrogen | A11122 RRID:AB_221569 | 1:1000 |
| Antibody | Anti-Brp (mouse monoclonal) | Developmental Studies Hybridoma Bank | nc82 RRID:AB_2341866 | 1:30 |
| Antibody | Anti-HA-Tag (mouse monoclonal) | Cell Signaling Technology | C29F4; #3724 RRID:AB_10693385 | 1:300 |
| Antibody | Anti-FLAG (rat monoclonal) | Novus Biologicals | NBP1-06712 RRID:AB_1625981 | 1:200 |
| Antibody | Anti-V5-TAG Dylight-549 (mouse monoclonal) | Bio-Rad | MCA2894D549GA RRID:AB_10845946 | 1:500 |
| Antibody | Anti-mouse IgG(H&L) AlexaFluor-568 (goat polyclonal) | Invitrogen | A11031 RRID:AB_144696 | 1:400 |
| Antibody | Anti-rabbit IgG(H&L) AlexaFluor-488 (goat polyclonal) | Invitrogen | A11034 RRID:AB_2576217 | 1:800 |
| Antibody | Anti-mouse IgG(H&L) AlexaFluor-488 conjugated (donkey polyclonal) | Jackson Immuno Research Labs | 715-545-151 RRID:AB_2341099 | 1:400 |
| Antibody | Anti-rabbit IgG(H&L) AlexaFluor-594 (donkey polyclonal) | Jackson Immuno Research Labs | 711-585-152 RRID:AB_2340621 | 1:500 |
| Antibody | Anti-rat IgG(H&L) AlexaFluor-647 (donkey polyclonal) | Jackson Immuno Research Labs | 712-605-153 RRID:AB_2340694 | 1:300 |
| Antibody | Anti-rabbit IgG(H+L) Alexa Fluor 568 (goat polyclonal) | Invitrogen | A-11036 RRID:AB_10563566 | 1:1000 |
| Chemical compound, drug | Pentyl acetate | Sigma-Aldrich | 109584 | 1:10,000 in paraffin oil |
| Chemical compound, drug | Ethyl lactate | Sigma-Aldrich | W244015 | 1:10,000 in paraffin oil |
| Chemical compound, drug | Paraffin oil | Sigma-Aldrich | 18512 | |
| Software, algorithm | ImageJ and Fiji | NIH; Schindelin et al., 2012 | https://imagej.nih.gov/ij/ http://fiji.sc/ | |
| Software, algorithm | MATLAB | MathWorks | https://www.mathworks.com/ | |
| Software, algorithm | Adobe Illustrator CC | Adobe Systems | https://www.adobe.com/products/illustrator.html | |
| Software, algorithm | GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | Caltech FlyTracker | Eyjolfsdottir et al., 2014 | https://github.com/kristinbranson/FlyTracker | |
| Software, algorithm | neuPrint | Plaza et al., 2022 | https://neuprint.janelia.org/ | |
| Software, algorithm | Cytoscape | Shannon et al., 2003 | https://cytoscape.org/ | |
| Software, algorithm | Janelia workstation | HHMI Janelia | https://doi.org/10.25378/janelia.8182256.v1 | |
| Software, algorithm | NeuTu | Zhao et al., 2018b; Zhao et al., 2018c | https://github.com/janelia-flyem/NeuTu | |
| Software, algorithm | VVD Viewer | Wan et al., 2012; Kawase et al., 2012 | https://github.com/takashi310/VVD_Viewer | |
| Other | Grade 3 MM Chr Blotting Paper | Whatmann | 3030-335 | Used in glass vials with paraffin oil diluted odors |
| Other | Mass flow controller | Alicat | MCW-200SCCM-D | Mass flow controller used for the olfactory arena |





