Human-induced pluripotent stem cell-derived microglia integrate into mouse retina and recapitulate features of endogenous microglia
Figures
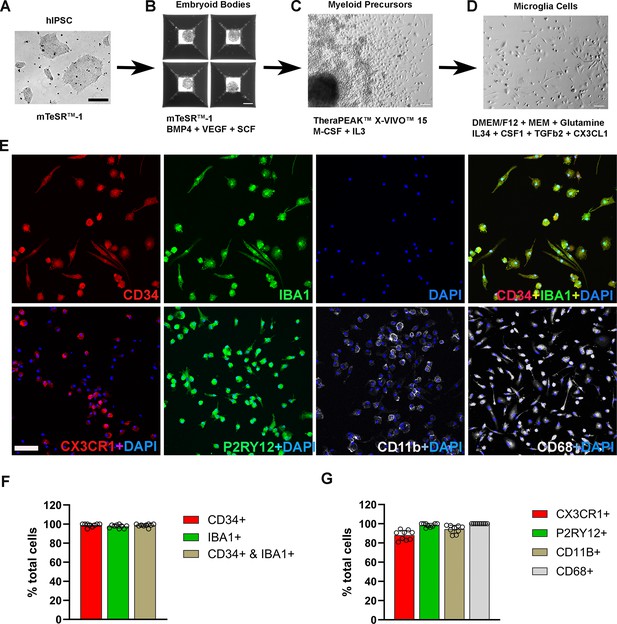
Differentiation and characterization of human-induced pluripotent stem cell (iPSC)-derived microglia.
(A) Human iPSCs were cultured in a 6-well plate. Scale bar = 200 µm. (B) Embryoid body formation was enabled in AggreWell800 plate at day 8 in culture medium mTeSR1 plus BMP4, VEGF, and SCF. Scale bar = 200 µm. (C) Image of a myeloid precursor cluster following 1 month culture of embryoid bodies in TheraPEAK X-vivo-15 Serum-free Hematopoietic Cell Medium with added M-CSF and IL3. Scale bar = 50 µm. (D) Image of microglial cells in maturation culture for 2 weeks with Dulbecco's Modified Eagle Medium (DMEM)/F12 plus non-essential amino acids, glutamine, IL34, CSF1, TGFb2, and CX3CL1. Scale bar = 50 µm. (E) Immunohistochemical staining for Iba1 and human CD34, CX3CR1, P2RY12, CD11b, and CD68. Scale bar = 100 µm. (F) Cell counts and colocalization analysis of (F) CD34- and Iba1-positive cells and (G) positivity for myeloid cell markers CX3CR1, CD11b, activation marker CD68, and microglia marker P2RY12 in differentiated microglia.
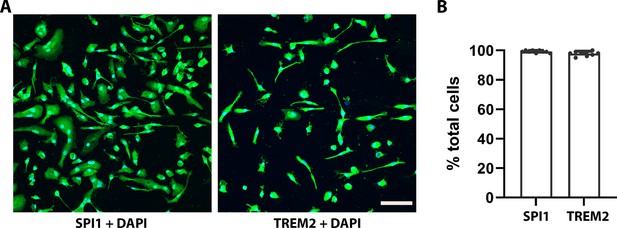
Immunocytochemistry staining with human SPI1 and TREM2.
(A) Confocal images showed SPI1 and TREM2 staining. Scale bar = 100 μm. (B) SPI1- and TREM2-positive cells are 99.7% and 99.4%, respectively, in entire DAPI+ cell counts.
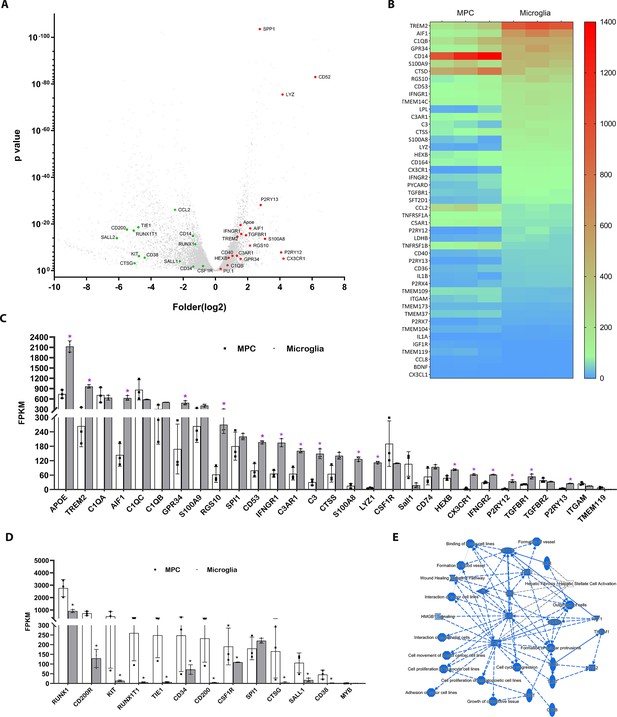
Profiling of genes differentially expression between differentiated microglial cells vs. myeloid progenitor cells (MPCs) using bulk RNAseq analysis.
(A) Volcano plot showing representative genes that were either upregulated (red) or downregulated (green) in differentiated microglia vs. MPCs. (B) Heat map showing increased expression of microglia-enriched genes in differentiated microglia (Supplementary file 1). (C) Histogram comparing the expression levels of microglia-enriched genes in terms of Fragments Per Kilobase of transcript per Million mapped reads (FPKM). *p < 0.05. (D) Histogram comparing expression levels of myeloid cell lineage genes in human-induced pluripotent stem cell (iPSC)-derived MPC and microglia cells using FPKM. *p < 0.05. (E) Graphic signaling pathway analysis with Ingenuity Pathway Analysis (IPA) highlighting IL6 and IL1B as signaling hubs in differential gene expression patterns (Supplementary file 1) in differentiated microglia vs. MPCs.
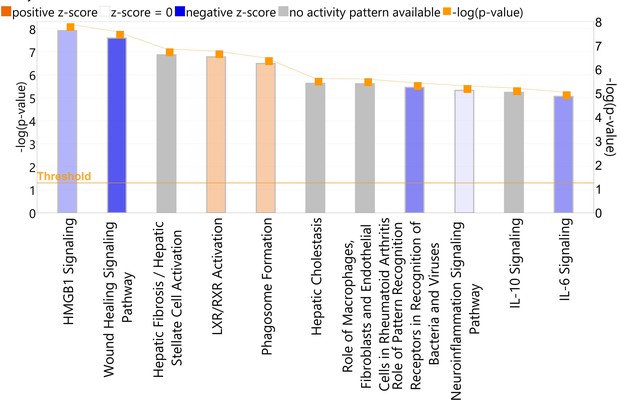
Analysis results of the canonical pathway of complete differentiated human-induced pluripotent stem cell (hiPSC)-derived microglia vs. myeloid progenitor cells with Ingenuity Pathway Analysis (IPA) with the enriched microglia genes (Supplementary file 1).

Hierarchical cluster analysis on microglia-enriched genes among human-induced pluripotent stem cell (hiPSC)-derived microglial cells (hiPSC-MG) and human adult brain microglia cells (AMG), fetal brain microglia cells (FMG), inflammatory monocytes (IM), monocytes (M).
The human microglia gene panel combined our mouse microglia-enriched genes and human microglia-enriched genes (Abud et al., 2017; Muffat et al., 2016; Douvaras et al., 2017; Böttcher et al., 2019; van der Poel et al., 2019). The total microglia-enriched gene list contains 203 genes (Supplementary file 2), which used to be extracted from the gene profile of human-induced pluripotent stem cell (hiPSC)-MG and downloaded human adult microglia (AMG), fetal microglia (FMG), inflammatory monocyte (IM) and monocytes (M) (GSE 178846, Abud et al., 2017). 188 genes (Supplementary file 2) were obtained from both gene lists. All the gene counts were normalized with four human cells housekeeping genes C1orf43, RAB7A, REEP5, and VCP (Eisenberg and Levanon, 2013). The hierarchical cluster was analyzed by JMP (JMP Statistical Discovery LLC). Results showed that hiPSC-MG is more comparable with AMG and FMG.
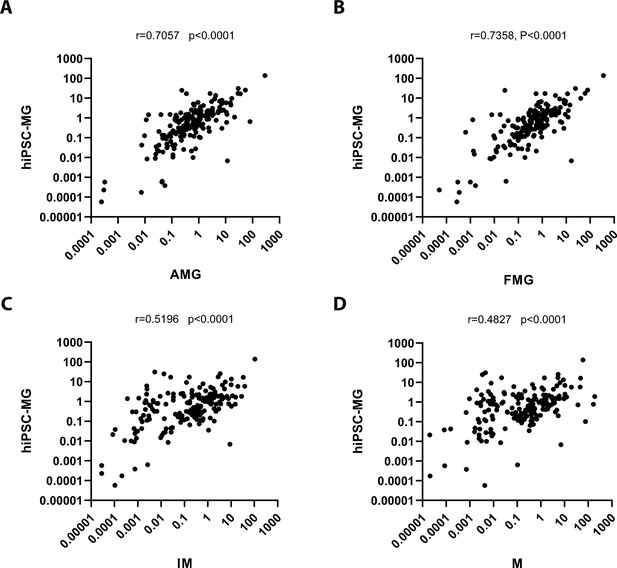
Correlation analysis between the expression levels of microglia-enriched genes in human-induced pluripotent stem cell (hiPSC)-derived microglia cells (hiPSC-MG) vs. those in other myeloid cells.
The correlation analysis of 188 human microglia-enriched genes (Supplementary file 2) among human-induced pluripotent stem cell (hiPSC)-derived microglia cells (hiPSC-MG), human adult brain microglia (AMG) cells (A), fetal brain microglia (FMG) cells (B), inflammatory monocytes (IM) (C), and monocytes (M) (D) (GSE 178846, Abud et al., 2017), respectively. The images and the analysis results (Prism, GraphPad) showed that expression levels of microglia-enriched genes in hiPSC-MG are more correlated between those in FMG (r = 0.7358, p < 0.0001) and AMG (r = 0.7057, p < 0.0001).
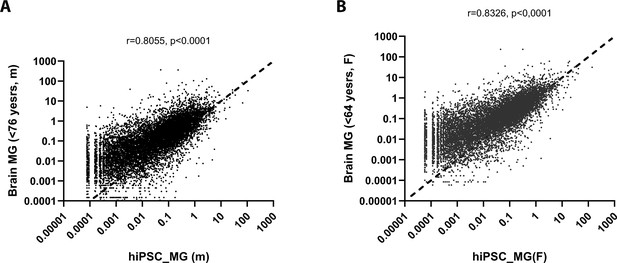
Correlation analysis between the expression levels of genes in human-induced pluripotent stem cell (hiPSC)-derived microglia cells (hiPSC-MG) vs. human donor brain microglia by sex.
Correlation comparison of entire gene profiles between male (A)/female (B) human-induced pluripotent stem cell (hiPSC)-derived microglial cells and male (A)/female (B) human brain microglia cells (GSE 111972, van der Poel et al., 2019; Supplementary file 3), respectively. The results showed they are significantly correlative (r = 0.8055 (M), r = 0.8326 (F), p < 0.0001).
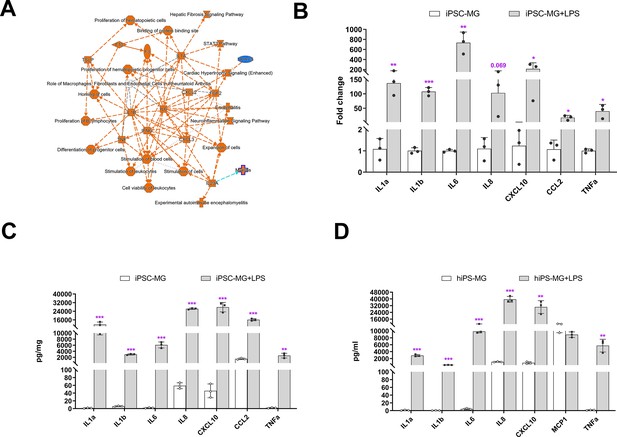
Inflammation responses of human-induced pluripotent stem cell (hiPSC)-derived microglial cells following lipopolysaccharide (LPS) stimulation.
(A) Ingenuity Pathway Analysis (IPA) showed different gene expression (fold change >twofold, p < 0.05) between LPS-treated and control hiPSC-derived microglial cells, demonstrating activation of core pathways involving IL6, IL1A, IL1B, and IFNG. (B) Assessment of mRNA expression of selected genes for inflammatory cytokines using quantitative reverse transcription-PCR (qRT-PCR; Oligonucleotide primers are provided in Supplementary 4) demonstrated increased expression following LPS (0.1 µg/ml) stimulation for 6 hr (3-6 replicates). These changes corresponded to increases in the protein expression levels of inflammatory cytokines following 24 hr of LPS stimulation as measured with a Multiplex kit (Millipore) in cell lysate (C) and conditioned media (D). The data in (C) and (D) are presented as means ± SEM (3-6 replicates). *p < 0.05, **p < 0.01, ***p < 0.001.
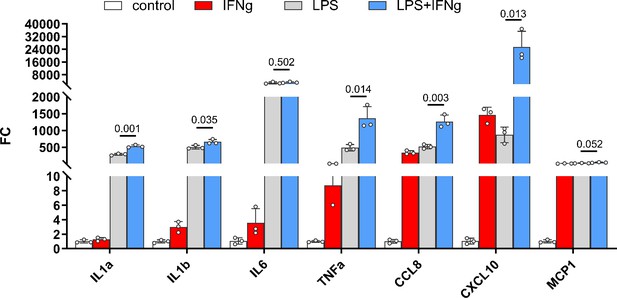
Inflammatory cytokines were produced synthetically by IFNG and LPS in human-induced pluripotent stem cell (hiPSC)-derived microglial cells.
hiPSC-derived microglial cells were cultured in 6-well plates for 14 days, 20 ng/ml of human IFNG and 0.5 µg/ml of LPS were added to the wells, respectively, or 20 ng/ml of human IFNG and 0.5 µg/ml of LPS were added to the wells together. After 6 hr of incubation, the cells were washed and harvested into a 1.5-ml Eppendorf tube for RNA isolation with a NucleoSpin RNA/Protein Mini kit (Macherey-Nagel, #740933.50). The cDNA was synthesized with PrimeScript 1st strand cDNA Synthesis Kit (Takara, #6110A). Quantitative reverse transcription-PCR (qRT-PCR) was performed using a SYBR green RT-PCR kit (Affymetrix), using the Bio-Rad CFX96 Touch Real-Time PCR Detection System under the following conditions: denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, and then 60°C for 45 s. Threshold cycle (CT) values were calculated and expressed as fold-induction determined using the comparative CT (2ΔΔCT) method. Ribosomal protein S13 (RPS13) and GAPDH were used as internal controls. Oligonucleotide primers are provided in Supplementary 4, the samples have 3 replicates. .
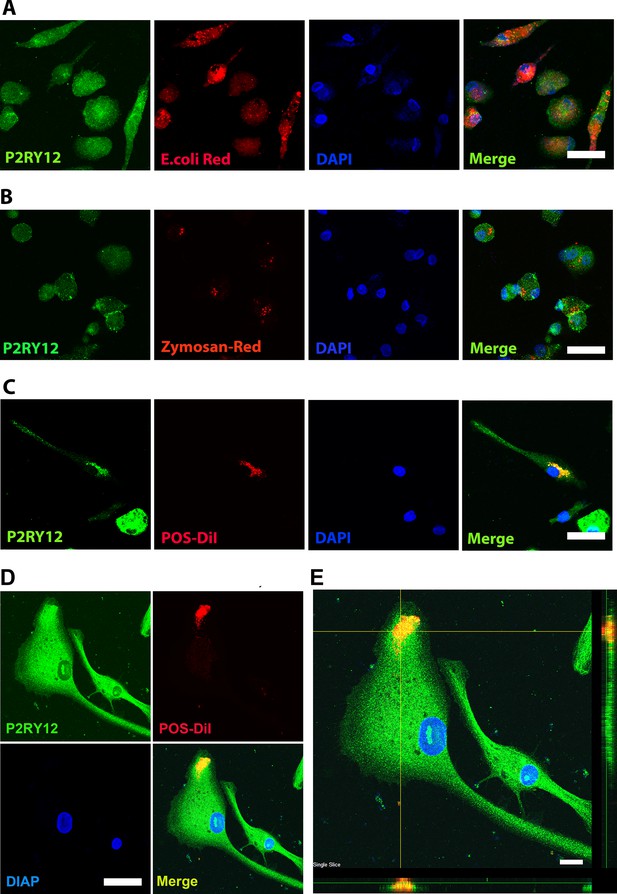
Human-induced pluripotent stem cell (iPSC)-derived microglia demonstrate robust phagocytosis.
Human iPSC-derived microglia were incubated for 1 hr in pHrodo Red E. coli bioparticles (A), pHrodo Red zymosan bioparticles (B), DiI-labeled bovine photoreceptor outer segments (POSs) (C) and labeled with anti-human P2RY12 antibody (green) and DAPI. Scale bar = 40 µm. (D) A high-magnification view of a POS-containing intracellular vesicle within a labeled microglial cell is shown. Scale bar = 40 µm. (E) An overlay of panels in (D) with side views. Scale bar = 40µm.

Floating myeloid progenitor cells were cultured in a 2-well slide chamber overnight, and then the DiI-labeled bovine photoreceptor outer segments were added to the chamber and incubated for 1 hr.
The cells were fixed with 4% paraformaldehyde (PFA) for 20 min and stained with IBA1 and hP2RY12, respectively. The DiI-labeled bovine photoreceptor outer segments can only be engulfed by IBA1- or P2RY12-positive microglia cells. Scale bar = 16 µm.
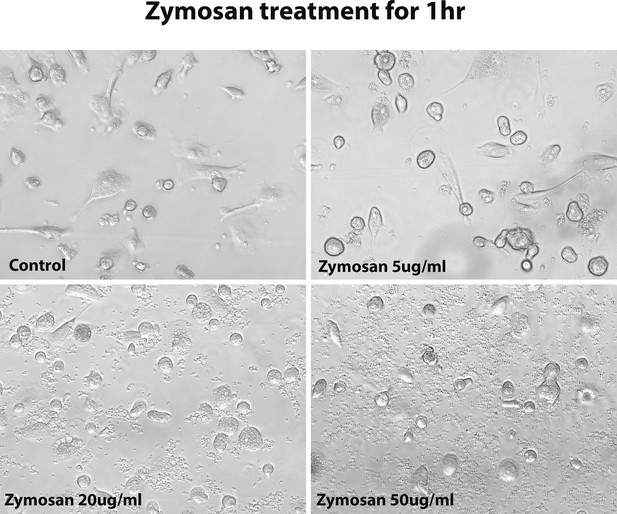
The morphology of human-induced pluripotent stem cell (hiPSC)-derived microglia cells under 1 hr zymosan treatment with different concentrations.
After 1 hr of treatment with zymosan, 5 µg/ml concentration did not change the microglia cell morphology, but over 20 µg/ml concentration changed the microglia morphology to an ameboid round shape.
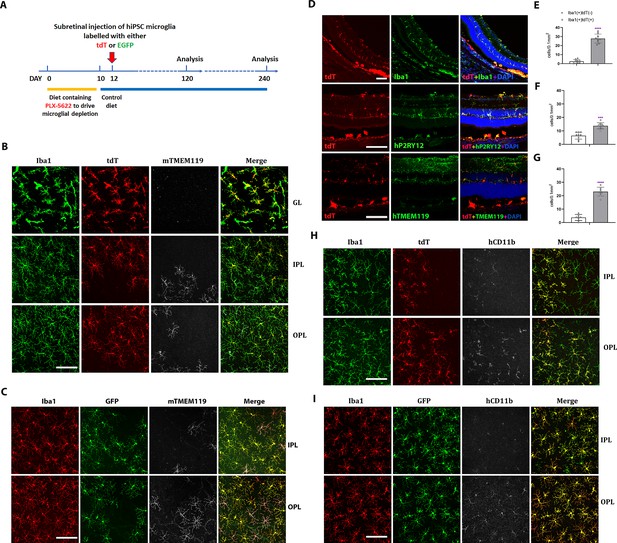
Xenotransplanted human-induced pluripotent stem cell (iPSC)-derived microglial cells into recipient mouse retina in vivo demonstrate recapitulation of endogenous distribution, cellular morphology, and stable integration for up to 4 months.
(A) The schematic diagram shows the timeline for transplantation experiments. Two-month-old adult transgenic Rag2−/−;IL2rg−/−;hCSF1+/+ mice were fed a PLX-5622-containing diet for 10 days before switching to standard chow. Two days following the resumption of standard chow, human iPSC-derived microglial cells expressing either tdTomato or EGFP were xenotransplanted into the subretinal space via subretinal injection (5000 cells in 1 µl injection volume). Retinas were harvested for analysis 120 and 240 days following transplantation. (B, C) The retinas isolated from post-transplantation were analyzed in flat-mounted tissue with confocal imaging. Transplanted human-induced pluripotent stem cell (hiPSC)-derived microglia were visualized through their expression of tdtomato (TdT) (B) or EGFP (C), while endogenous mouse microglia were visualized using immunostaining for mouse Tmem119 (mTmem119). Imaging analysis was performed in separate layers of the retina, including the ganglion cell layer (GL), inner plexiform layer (IPL), and outer plexiform layer (OPL). Scale bar = 100 µm. (D) The retinal section showed human iPSC-derived microglial cells integrated into whole retinal layers (top panel) and positively stained with human P2RY12 and TMEM119 microglia signature markers. Scale bar = 100 µm. The microglia cell number in GL, IPL, and OPL of host mouse retina were counted: mouse microglial cells (Iba1+, tdT−) and grafted human microglial cells (Iba1+, tdT+) were shown in (E), (F), and (G), respectively. ***p < 0.001, ****p < 0.0001, 3-6 biological replicates were performed. (H) and (I) showed tdT (H) or EGFP (I) labeled human iPSC-derived microglial cells in the IPL and OPL of the flat-mount retina with human CD11b staining. These results demonstrated that the infiltration of grafted hiPSC-derived microglial cells integrated into the mouse retina is general in nature and not cell line specific. Scale bar = 100 µm.

Homeostatic human-induced pluripotent stem cell (hiPSC)-derived microglial cells in the mouse retina do not affect local retinal cells.
Entire section images showed GFAP, GS, RBPMS, and Iba1 staining for astrocytes, Müller cells, ganglion cells, and microglia cells in the retina after 4 months of xenotransplantation. Scale bar = 300 µm.
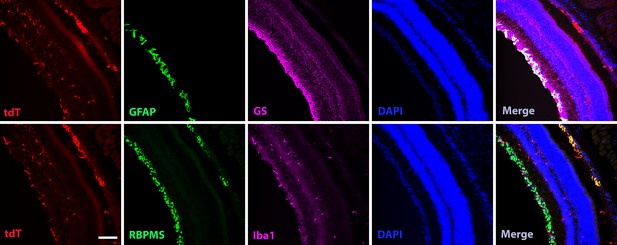
Homeostatic human-induced pluripotent stem cell (iPSC)-derived microglia cells in the mouse retina do not affect local retinal cells.
The high-magnification images showed GFAP, GS, RBPMS, and Iba1 staining for astrocytes, Müller cells, ganglion cells, and microglia cells in the retina after 4 months of transplantation. Scale bar = 100 µm.
Homeostatic human-induced pluripotent stem cell (iPSC)-derived microglia cells in the mouse retina do not affect local retinal cells.
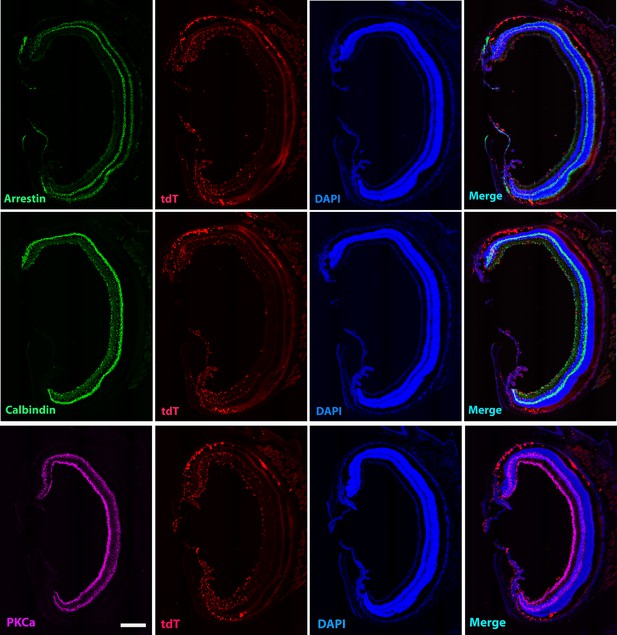
Entire section images showed arrestin, calbindin, and PKCα staining for cone photoreceptors, horizontal and some amacrine cells, and bipolar cells in the retina after 4 months of xenotransplantation. Scale bar = 300 µm.

Homeostatic human-induced pluripotent stem cell (iPSC)-derived microglia cells in the mouse retina do not affect local retinal cells.
The partial section of high-magnification images showed arrestin, calbindin, and PKCα staining for cone photoreceptors, horizontal and some amacrine cells, and bipolar cells in the retina after 4 months of xenotransplantation. Scale bar = 100 µm.

Homeostatic human-induced pluripotent stem cell (iPSC)-derived microglia cells in the mouse retina do not take over local retinal microglia cells.
The section images showed Red Fluorescence Protein (RFP) and mouse CD11b staining to determine the tdT+ human microglia cells and local mouse microglia cells in the retina after 4 months of xenotransplantation. The results showed that the tdT+ human microglia cells colocalized with RFP staining (Far red) but not mouse CD11b (green) staining. Scale bar = 100 µm.
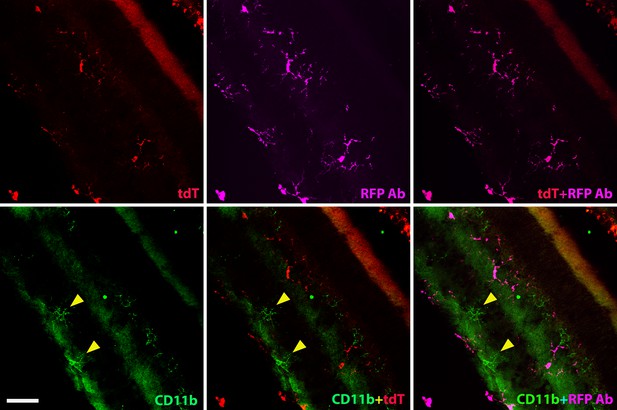
Homeostatic human-induced pluripotent stem cell (iPSC)-derived microglia cells in the mouse retina do not take over local retinal microglia cells.
The hight magnification images showed RFP and mouse CD11b staining in the retina after 4 months of xenotransplantation. The results showed that the tdT+ human cells colocalized with RFP staining (Far red) but not mouse CD11b (green) staining. The triangle marker indicated that the local mouse microglia cells were only stained with mouse CD11b but not colocalized with tdT+ human microglia cells. Scale bar = 50 µm.
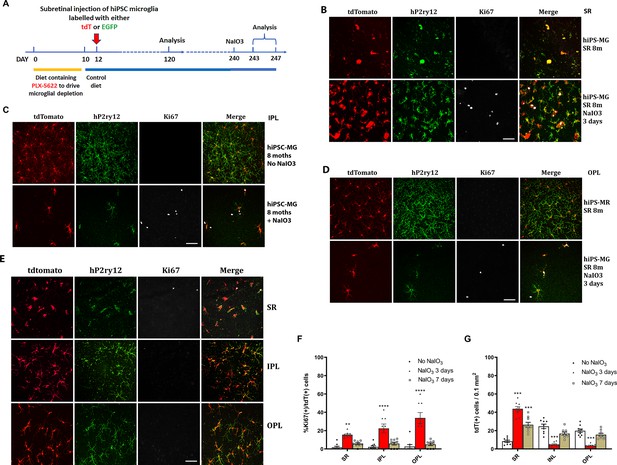
Migration and proliferation of human-induced pluripotent stem cell (hiPSC)-derived microglia in the mouse retina after sodium iodate (NaIO3)-induced retinal pigment epithelial (RPE) cell injury.
(A) The schematic diagram shows the experiment’s procedure. After 8 months post-transplantation of hiPSC-derived microglia, recipient animals were administered NaIO3 (30 mg/kg body weight, intraperitoneal injection) to induce RPE injury. Retinas were harvested at 3 and 7 days after NaIO3 administration and microglia numbers in the retina and subretinal space will be monitored in retina and RPE-choroid flat mounts. (B) RPE-choroid flat mounts demonstrate an increase of hiPSC-derived microglia (tdTomato+ and P2RY12+) in the subretinal space in response to RPE injury. A subset of subretinal microglia labeled for Ki67 indicates active proliferation. Scale bar = 60 µm. (C) and (D) showed the number of P2RY12+ and tdtomato+ human microglial cells in inner plexiform layer (IPL) (C) and outer plexiform layer (OPL) (D) decreased; some of them showed Ki67+ staining, Scale bar = 60 µm. The cell count results showed in (F) and (G). (E) The retinal flat mount showed the number of P2RY12+ and tdtomato+ human microglial cells in IPL and OPL that were repopulated, and the cells stopped dividing with loss of the Ki67 staining at 7 days after NaIO3 injection. The cell numbers are shown in (F) and (G) (3 biological replicates). Scale bar = 60 µm, ** P<0.01, *** P<0.001, **** P,0.0001.

The images of hP2RY12 staining on the retina after 8 months of xenotransplantation.
The images showed the tdtomato+ human microglia cells colocalized with hP2RY12 staining in GL, inner plexiform layer (IPL), and outer plexiform layer (OPL) in the mouse retina. Scale bar = 300 µm.

The images of hTMEM119 staining on the retina after 8 months of xenotransplantation.
The images showed the tdtomato+ human microglia cells colocalized with hTMEM119 staining in GL, inner plexiform layer (IPL), and outer plexiform layer (OPL) in the mouse retina. Scale bar = 300 µm.
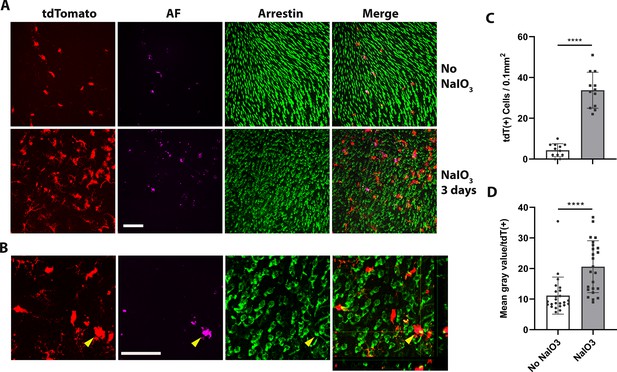
Dyshomeostatic human-induced pluripotent stem cell (iPSC)-derived microglial cells in the mouse retina phagocytose dead photoreceptor cells/debris after retinal pigment epithelial (RPE) cell injury.
(A) Dyshomeostatic human microglial cells (tdtomato+) accumulated in the photoreceptor cell layer after 3 days of sodium iodate (NaIO3)-induced RPE cell injury compared with no NaIO3 administration. The photoreceptor cells stained with cone arrestin (green) and autofluorescence showed in magenta. Scale bar = 60 µm. (B) High-magnificent images and the side view showed human microglial cells (red) co-labeled with photoreceptor cells arrestin staining (green) after 3 days of NaIO3 injury. The yellow arrowhead showed the colocalized tdT+ human microglia cell and arrestin+ cone photoreceptor cell. Scale bar = 40 µm. (C) The number of tdtomato+ human microglial cells in the photoreceptor layer. (D) The mean gray autofluorescence value in each human microglia cell. ****p < 0.0001.
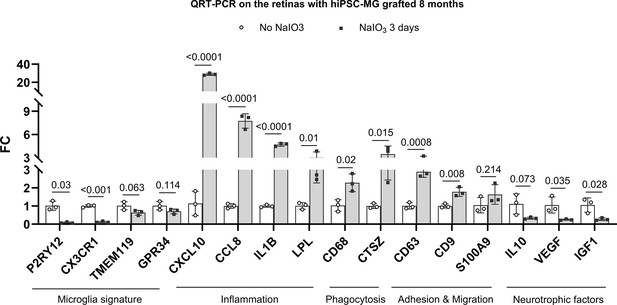
The inflammation, phagocytosis, adhesion and migration, neurotrophic factors, and microglia signature gene expression in human-induced pluripotent stem cell (hiPSC)-derived microglia cells of grafted retinas.
The gene coding sequences were compared between humans and mice, a human-specific sequence was chose to make the oligos (Supplementary file 5), and ran a quantitative reverse transcription-PCR (qRT-PCR) on 8-month hiPSC-derived microglia cells grafted retinas with/without sodium iodate (NaIO3)-treated retinas. The results revealed that hiPSC-derived microglia cells expressed more inflammatory factors and phagocytosis genes and promoted cell migration but decreased microglia cell signature genes and neurotrophic factors in NaIO3-treated retina (3 biological replicates in each group).
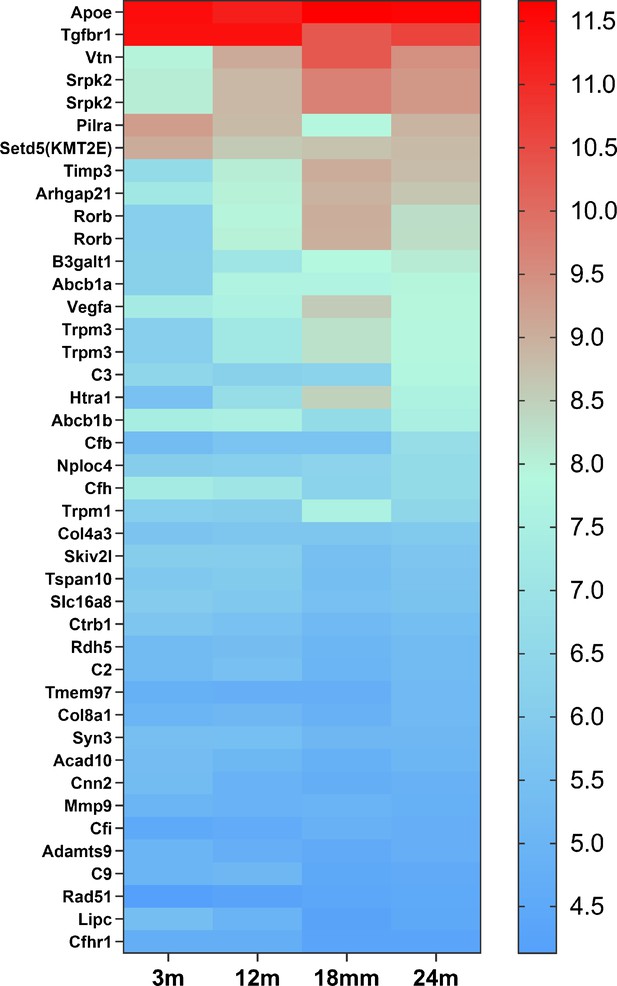
The heat map of 42 candidate genes from 34 loci associated with age-related macular degeneration (AMD) expressed in retinal microglia cells.
The microglia gene expression data are from microarray data previously published (Ma et al., 2013). The candidate genes came from the published paper (den Hollander et al., 2022). The gene list is in Supplementary file 6.
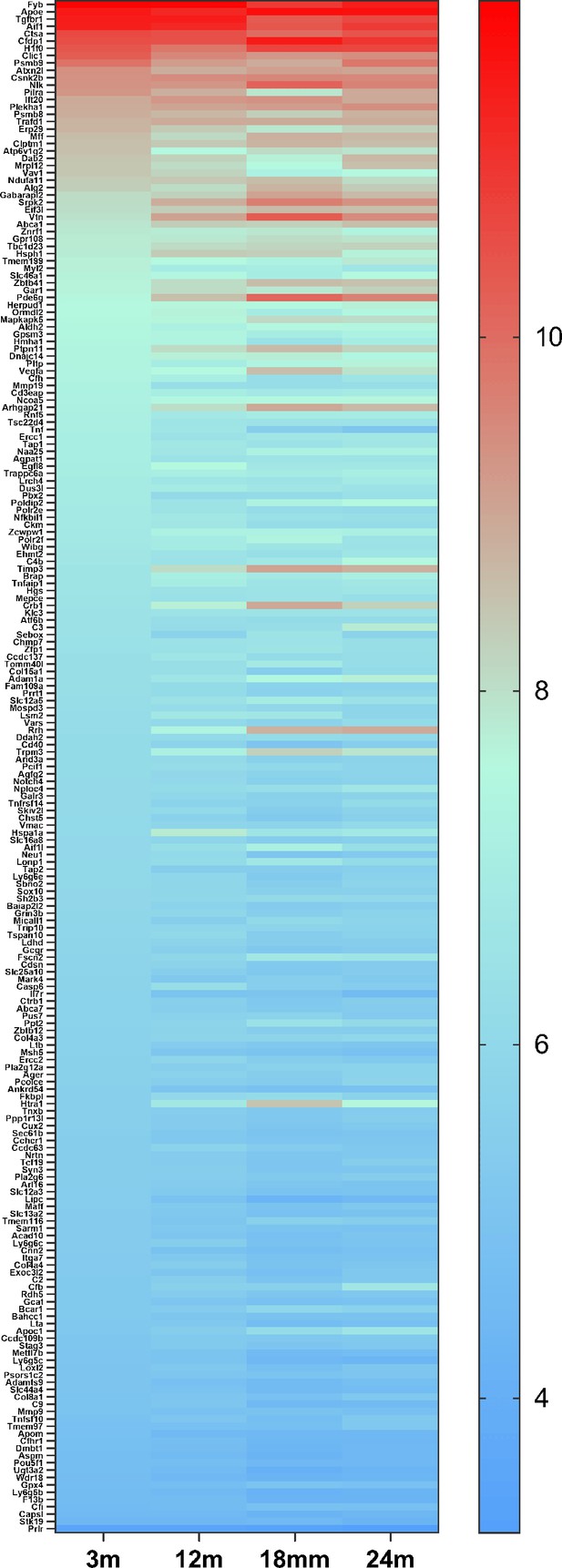
The heat map of 209 genes associated with age-related macular degeneration (AMD) (Fritsche et al., 2016) expressed in retinal microglia cells.
The microglia gene expression data are from microarray data previously published (Ma et al., 2013). The gene list is in Supplementary file 7.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | C;129S4-Rag2tm1.1Flv Csf1tm1(CSF1)Flv Il2rgtm1.1Flv/J | PubMed:21791433 | MGI:J:177073 | RRID:IMSR_JAX:017708 |
| Genetic reagent (Homo sapiens) | KYOUDXR0109B | ATCC | ACS-1023 | Human-induced pluripotent stem cells (iPSCs) |
| Genetic reagent (Homo sapiens) | NCRM6 | NHLBI | NCRM6 (female) iPSC line | From CD34+ cells, Episomal vectors |
| Genetic reagent (Homo sapiens) | MS19-ES-H | NHLBI | MS19-ES-H (female) iPSC line | From PBMS cells, Cytotune Sendai Virus kit |
| Genetic reagent (Homo sapiens) | NCRM5-AAVS1-CAG-EGFP | NHLBI | NCRM5-AAVS1-CAG-EGFP (clone 5) | From CD34+ cells, NCRM5 (male) reporter iPSC line with CAG-EGFP targeted mono-allelically at AAVS1 safe harbor |
| Genetic reagent (Homo sapiens) | ND2-AAVS1-iCAG-tdTomato | NHLBI | ND2-AAVS1-iCAG-tdTomato (clone 1) | From fibroblast cells, ND2 (male) reporter iPSC line with insulated CAG-tdTomato targeted mono-allelically at AAVS1 safe harbor |
| Antibody | anti-Iba1 (rabbit polyclonal) | Wako | Cat. #: 019-19741, RRID:AB_839504 | IHC (1:500) |
| Antibody | anti-human TMEM119 (rabbit polyclonal) | Sigma-Aldrich | Cat. #: HPA051870, RRID:AB_2681645 | IHC (1:100) |
| Antibody | anti-human CD68 (mouse monoclonal) | R&D | Cat. #: MAB20401 | IHC (1:100) |
| Antibody | anti-human CD45 (mouse monoclonal) | R&D | Cat. #: FAB1430R | IHC (1:100) |
| Antibody | anti-human CD11b (mouse monoclonal) | R&D | Cat. #: FAB1699R | IHC (1:100) |
| Antibody | anti-human CX3CR1 (rat monoclonal) | Invitrogen | Cat. #: 61-6099-42 | IHC (1:100) |
| Antibody | anti-human HLA (mouse monoclonal) | Invitrogen | Cat. #: 11-9983-42 | IHC (1:100) |
| Antibody | anti-mouse CD11b (rat monoclonal) | Bio-Rad | Cat. #: MCA711G | IHC (1:100) |
| Antibody | anti-GFAP (rat monoclonal) | Invitrogen | Cat. #: 13-0300, RRID:AB_2532994 | IHC (1:200) |
| Antibody | anti-mouse TMEM119 (guinea pig polyclonal) | Synaptic systems | Cat. #: 400 004, RRID:AB_2832239 | IHC (1:500) |
| Antibody | anti-CD68 (rat monoclonal) | Bio-Rad | Cat. #: MCA1957, RRID:AB_322219 | IHC (1:200) |
| Antibody | anti-CD34 (rat monoclonal) | eBioscience | Cat. #: 14-0341 | IHC (1:30) |
| Antibody | anti-PU.1 (rabbit monoclonal) | Thermo Fisher Scientific | Cat. #: MA5-15064 | IHC (1:200) |
| Antibody | anti-Trem2 (rabbit monoclonal) | Thermo Fisher Scientific | Cat. #: 702886 | IHC (1:100) |
| Antibody | anti-CD45 (rat monoclonal) | Bio-Rad | Cat. #: MCA1388, RRID:AB_321729 | IHC (1:100) |
| Antibody | anti glutamine synthetase (mouse monoclonal) | Millipore | Cat. #: MAB302, RRID:AB_2110656 | IHC (1:200) |
| Antibody | anti-RBPMS (guinea Pig polyclonal Ab) | Phosphosolutions | Cat. #: 1832-RBPMS | IHC (1:100) |
| Antibody | anti-cone arrestin (rabbit polyclonal) | Millipore | Cat. #: AB15282, RRID:AB_1163387 | IHC (1:200) |
| Antibody | anti-calbindin (rabbit polyclonal) | Swant | Cat. #: CB-38a | IHC (1:5000) |
| Antibody | anti-PKCa (rabbit polyclonal) | Sigma-Aldrich | Cat. #: P4334, RRID:AB_477345 | IHC (1:200) |
| Antibody | anti-RFP (rabbit polyclonal) | RockLand | Cat. #: 600-401-379-RTU | IHC (1:100) |
| Antibody | anti-Ki67-660 (rat monoclonal) | eBioscience | Cat. #: 50-5698-82, RRID:AB_2574235 | IHC (1:50) |
| Antibody | anti-P2RY12 (rabbit polyclonal) | Thermo Fisher Scientific | Cat. #: PA5-77671, RRID:AB_2736305 | IHC (1:100) |
| Antibody | anti-P2RY12 (rabbit polyclonal) | Sigma-Aldrich | Cat. #: HPA014518, RRID:AB_2669027 | IHC (1:100) |
| Antibody | Goat anti-Rabbit IgG Alexa Fluor 488 | Invitrogen | Cat. #: A27034, RRID:AB_2536097 | IHC (1:200) |
| Antibody | Goat anti-Rabbit IgG Alexa Fluor 568 | Invitrogen | Cat. #: A11011, RRID:AB_143157 | IHC (1:200) |
| Antibody | Goat anti-Rabbit IgG Alexa Fluor 647 | Invitrogen | Cat. #: A32733, RRID:AB_2633282 | IHC (1:200) |
| Antibody | Goat anti-mouse IgG Alexa Fluor 488 | Invitrogen | Cat. #: A28175, RRID:AB_2536161 | IHC (1:200) |
| Antibody | Goat anti-mouse IgG Alexa Fluor 568 | Invitrogen | Cat. #: A-11004, RRID:AB_141371 | IHC (1:200) |
| Antibody | Goat anti-mouse IgG Alexa Fluor 647 | Invitrogen | Cat. #: A-21235, RRID:AB_141693 | IHC (1:200) |
| Antibody | Donkey anti-Rat IgG Alexa Fluor 488 | Invitrogen | Cat. #: A-21208, RRID:AB_141709 | IHC (1:200) |
| Antibody | Donkey anti-Rat IgG Alexa Fluor 594 | Invitrogen | Cat. #: A-21209, RRID:AB_2535795 | IHC (1:200) |
| Antibody | Donkey anti-Rat IgG Alexa Fluor 650 | Invitrogen | Cat. #: SA5-10029, RRID:AB_2556609 | IHC (1:200) |
| Antibody | Rat monoclonal anti CD11b, Alexa Fluor 488 | eBioscience | Cat. #: 53-0112-82, RRID:AB_469901 | IHC (1:50) |
| Peptide, recombinant protein | human M-CSF | Invitrogen | Cat. #: PHC9501 | |
| Peptide, recombinant protein | Human IL3 | R&D | Cat. #: 203-IL-100 | |
| Peptide, recombinant protein | human IL-34 | Peprotech | Cat. #: 200–34 | |
| Peptide, recombinant protein | human CX3CL1 | Peprotech | Cat. #: 300–31 | |
| Peptide, recombinant protein | human TGFb1 | R&D | Cat. #: 7666-MB-005 | |
| Peptide, recombinant protein | human TGFb2 | R&D | Cat. #: 7346-B2-005 | |
| Peptide, recombinant protein | human BMP-4 | GIBCO | Cat. #: PHC9534 | |
| Peptide, recombinant protein | human SCF | Miltenyi Biotec | Cat. #: 130096692 | |
| Peptide, recombinant protein | human VEGF | GIBCO | Cat. #: PHC9394 | |
| Chemical compound | PLX5622 | Plexxikon | PLX5622 was provided by Plexxikon Inc and formulated in AIN-76A standard chow by Research Diets Inc | 1200 mg/kg in chow |
| Chemical compound | NaIO3 | Sigma-Aldrich | Cat. #: S4007 | |
| Chemical compound | BSA | Sigma-Aldrich | Cat. #: A2153 | |
| Chemical compound | FBS | Thermo Fisher Scientific | Cat. #: A3160702 | |
| Chemical compound | Ketamine | Anased | Cat. #: NDC13985-584-10 | |
| Chemical compound | Xylazine | Anased | Cat. #: NDC59399-110-20 | |
| Chemical compound | Topical tropicamide | Alcon | Cat. # 215340 | |
| Chemical compound | Phenylephrine | Alcon | Cat. l# 215664 | |
| Chemical compound | 0.5% Proparacaine HCL | Sandoz | Cat. #: 101571 | |
| Chemical compound | Surcrose | Sigma-Aldrich | Cat. #: S7903-5KG | |
| Chemical compound | OCT | Thermo Fisher Scientific | Cat. #: 23-730-571 | |
| Chemical compound | Fluorescein AK-FLUOR | Akorn | Cat. #: 17478-253-10 | |
| Chemical compound | Tamoxifen | Sigma-Aldrich | Cat. #: T5648-5G | |
| Chemical compound | HBSS | Sigma-Aldrich | Cat. #: H8264-1L | |
| Chemical compound | L-(+)-Cysteine hydrochloride monohydrate | Fisher | Cat. #: C562-25 | |
| Chemical compound | Papain, lyophilized | Worthington Biochemical | Cat. #: LS003119 | |
| Chemical compound | DNAse I | Worthington Biochemical | Cat. #: LS006333 | |
| Chemical compound | Superoxide dismutase | Worthington Biochemical | Cat. #: LS003540 | |
| Chemical compound | Catalase | Sigma-Aldrich | Cat. #: C1345-1G | |
| Chemical compound | (+)-α-Tocopherol acetate | Sigma-Aldrich | Cat. #: T-1157-1G | |
| Chemical compound | Gentamicin solution | Sigma-Aldrich | Cat. #: G1397-10ml | |
| Chemical compound | D-(+)-Glucose | Sigma-Aldrich | Cat. #: G7021-100g | |
| Chemical compound | Antipain dihydrochloride | Roche | Cat. #: 11004646001 | |
| Chemical compound | HEPES | Invitrogen | Cat. #: 15630080 | |
| Chemical compound | EDTA | KD medical | Cat. #: RGC-3130 | |
| Chemical compound | RNAlater solution | Ambion | Cat. #: AM7021 | |
| Chemical compound | Triton X-100 | Sigma-Aldrich | Cat. #: X100100ml | |
| Chemical compound | Tween 20 | Sigma-Aldrich | Cat. #: P1379-100ml | |
| Chemical compound | Paraformadehyde | Fisher Scientific | Cat. #: 50-259-97 | |
| Chemical compound | Donkey serum | Sigma-Aldrich | Cat. #: D9663-10ml | |
| Chemical compound | Goat serum | Sigma-Aldrich | Cat. #: G9023-10ml | |
| Commercial assay or kit | Bloking Reagent | Sigma-Aldrich | Cat. #: 11096176001 | |
| Commercial assay or kit | In Situ Cell Death Detection Kit, TMR red | Sigma-Aldrich | Cat. #: 12156792910 | |
| Commercial assay or kit | Ib4 Alexa Fluor 568 | Invitrogen | Cat. #: I21412 | IHC (1:200) |
| Commercial assay or kit | Ib4 Alexa Fluor 647 | Invitrogen | Cat. #: I32450 | IHC (1:200) |
| Commercial assay or kit | DAPI | Sigma-Aldrich | Cat. #: D9542 | IHC (1:200) |
| Commercial assay or kit | Mounting medium without DAPI | Vector | Cat. #: H-1000 | |
| Commercial assay or kit | Mounting medium with DAPI | Vector | Cat. #: H-1200 | |
| Commercial assay or kit | RNeasy Mini Kit | QIAGEN | Cat. #: 74104 | |
| Commercial assay or kit | Rnase free Dnase set | QIAGEN | Cat. #: 79254 | |
| Commercial assay or kit | First strand cDNA synthesis | Takara | Cat. #: 6110A | |
| Commercial assay or kit | MessageBooster cDNA synthesis kit | Epicentre | Cat. #: MB060110 | |
| Commercial assay or kit | Fast SYBR Green Master Mix | Thermo Fisher Scientific | Cat. #: 4385617 | |
| Commercial assay or kit | LiDirect Lightening genotyping kit | LifeSci | Cat. #: M0015 | |
| Commercial assay or kit | eBioscience Flow Cytometry Staining Buffer | Thermo Fisher Scientific | Cat. #: 00-4222-57 | |
| Commercial assay or kit | X-VIVO-15 medium | Lonza | Cat. #: BEBP02-061Q | |
| Commercial assay or kit | DMEM:F12 medium | Thermo Fisher Scientific | Cat. #: 11330057 | |
| Commercial assay or kit | mTeSR1 | Stemcell technologies | Cat. #: 85850 | |
| Commercial assay or kit | N2 supplement | Thermo Fisher Scientific | Cat. #: 17502048 | |
| Commercial assay or kit | Non-essential Amino Acids (NEAA) | Thermo Fisher Scientific | Cat. #: 11140050 | |
| Commercial assay or kit | GlutaMax Supplement | Thermo Fisher Scientific | Cat. #: 35050061 | |
| Commercial assay or kit | Geltrex | Thermo Fisher Scientific | Cat. #: A1413301 | |
| Commercial assay or kit | TrypLE Express | Thermo Fisher Scientific | Cat. #: 12605010 | |
| Commercial assay or kit | Rho-kinase inhibitor Y-27632 | abcam | Cat. #: ab143784 | |
| Commercial assay or kit | mFreSR | Stemcell technologies | Cat. # 05854 | |
| Commercial assay or kit | Stem Cell Dissociation Reagent | ATCC | Cat, #: ACS-3010 | |
| Commercial assay or kit | Stem Cell Freezing Media | ATCC | Cat. #: ACS-3020 | |
| Commercial assay or kit | Penicillin-Streptomycin | Thermo Fisher Scientific | Cat. #: 15140122 | |
| Commercial assay or kit | pHrodo Red E. coli BioParticles | Thermo Fisher Scientific | Cat. #: P35361 | |
| Commercial assay or kit | pHrodo Red Zymosan Bioparticles | Thermo Fisher Scientific | Cat. #: P35364 | |
| Commercial assay or kit | Bovine rod outer segment | Invision Bioresources | Cat. #: 98740 | |
| Commercial assay or kit | Vybrant DiI Cell-Labeling Solution | Thermo Fisher Scientific | Cat. #: V22885 | |
| Commercial assay or kit | Lipopolysaccharides (LPS) | Sigma-Aldrich | Cat. #: L2630 | |
| Commercial assay or kit | RIPA lysis buffer | Sigma-Aldrich | Cat. #: R0278 | |
| Commercial assay or kit | proteinase inhibitor mixture | Calbiochem | Cat. #: 539132 | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat. #: 23227 | |
| Commercial assay or kit | Milliplex bead assay kit | Millipore | Cat. #: MCYTOMAG-70K | |
| Commercial assay or kit | AggreWellsTM800 | Stemcell technologies | Cat. #: 34825 | |
| Software | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software | GraphPad Prism7 | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 7 |
| Software | IPA | QIAGEN | RRID:SCR_008653 | |
| Software | JMP | JMP | RRID:SCR_014242 | Version 12 |
Additional files
-
Supplementary file 1
Microglia-enriched gene list.
The gene list of 71 microglia-enriched genes was extracted from bulk RNAseq of microglial cells vs. myeloid progenitor cells (MPCs). The total microglia-enriched gene list combined from the research conducted by Barres BA Lab (Bennett et al., Proc Natl Acad Sci U S A, 2016) and from our RNA sequencing data of mouse retinal microglia (Ma et al., 2013), identifying a total of 130 genes predominantly expressed in microglia.
- https://cdn.elifesciences.org/articles/90695/elife-90695-supp1-v1.xlsx
-
Supplementary file 2
188 hMG gene comparisons vs. GSE178846.
The human microglia gene panel combined our mouse microglia-enriched genes and human microglia-enriched genes (Abud et al., 2017; Muffat et al., 2016; Douvaras et al., 2017; Böttcher et al., 2019; van der Poel et al., 2019). The total microglia-enriched gene list contains 203 genes, which used to be extracted from the gene profile of hiPSC-MG and downloaded human adult microglia (AMG), fetal microglia (FMG), inflammatory monocyte (IM) and monocytes (M) (GSE 178846, Abud et al., 2017). 188 genes were obtained from both gene lists. All the gene counts were normalized with four human cells housekeeping genes C1orf43, RAB7A, REEP5, and VCP (Eisenberg and Levanon, 2013).
- https://cdn.elifesciences.org/articles/90695/elife-90695-supp2-v1.xls
-
Supplementary file 3
The gene list of comparison vs. GSE111972.
The total gene list of both male and female hiPSC-derived microglial cells and human brain microglia cells (GSE 111972, van der Poel et al., 2019).
- https://cdn.elifesciences.org/articles/90695/elife-90695-supp3-v1.xlsx
-
Supplementary file 4
The oligos for quantitative reverse transcription-PCR (qRT-PCR).
The oligos were used for qRT-PCR in cultured hiPSC-derived cells.
- https://cdn.elifesciences.org/articles/90695/elife-90695-supp4-v1.docx
-
Supplementary file 5
Human-specific oligos for quantitative reverse transcription-PCR (qRT-PCR).
The oligos were used for qRT-PCR in mouse retinas with integrated hiPSC-derived microglial cells.
- https://cdn.elifesciences.org/articles/90695/elife-90695-supp5-v1.xlsx
-
Supplementary file 6
42 genes associated with age-related macular degeneration (AMD).
42 candidate genes from 34 loci associated with AMD expressed in retinal microglia cells. The microglia gene expression data are from microarray data previously published (Ma et al., 2013). The candidate genes came from the published paper (den Hollander et al., 2022).
- https://cdn.elifesciences.org/articles/90695/elife-90695-supp6-v1.xlsx
-
Supplementary file 7
209 genes associated with age-related macular degeneration (AMD).
The 209 microglia-enriched genes were extracted from the genes associated with AMD (Fritsche et al., 2016). The microglia gene expression data are from microarray data previously published (Ma et al., 2013).
- https://cdn.elifesciences.org/articles/90695/elife-90695-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/90695/elife-90695-mdarchecklist1-v1.docx