Transformation of valence signaling in a mouse striatopallidal circuit
Figures
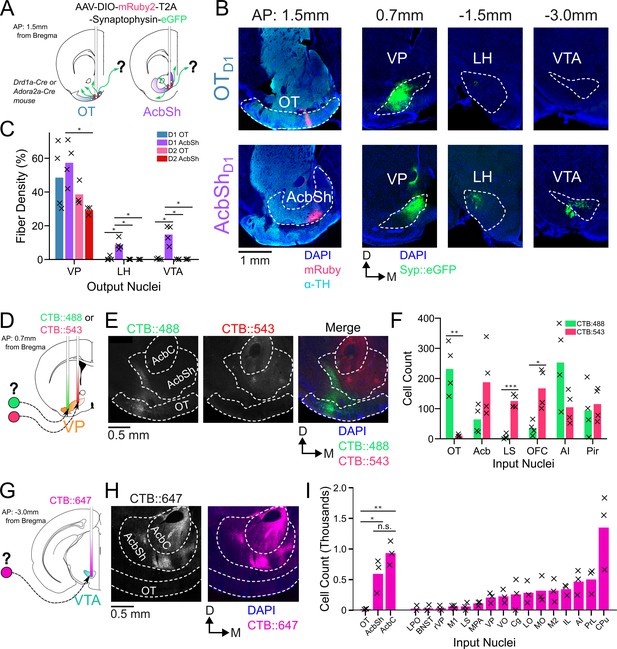
OTD1 and OTD2 primarily project to the lateral portion of the VP.
(A) Schematic representation of Cre-dependent anterograde axonal AAV tracing experiments used to characterize outputs of OT neurons. Drd1+ and Drd2+ neurons were separately labeled by using Drd1-Cre and Adora2a-Cre mouse lines, respectively. (B) Representative images from OTD1 (top) vs. the AcbShD1 injection (bot). Target sites (far-left column) are stained with ⍺-tyrosine hydroxylase antibodies to visualize the boundary between VP and OT. (C) Quantifying the % of output regions with fluorescence (n=3–4). (D) Schematic representation of two-color retrograde CTB tracing experiment used to confirm OT to VP connectivity. CTB::488 and CTB::543 were injected to the lateral and medial portion of the VP, respectively. (E) Representative images of CTB labeled neurons in the OT and Acb. (F) The number of labeled cells was quantified (n=4). (G) Schematic representation of retrograde CTB tracing experiment used to test OT to VTA connectivity. CTB::647 was injected in the VTA. (H) Representative image shows robust AcbSh and AcbC labeling but no OT labeling. (I) Quantification of labeling in different nuclei (n=3). Pairwise comparisons were done using the Student’s t-test. The p-values were corrected for FDR by Benjamini-Hocherg procedure. ***p<0.001, **p<0.01, *p<0.05. See Appendix 1—tables 1–3 for detailed statistics.
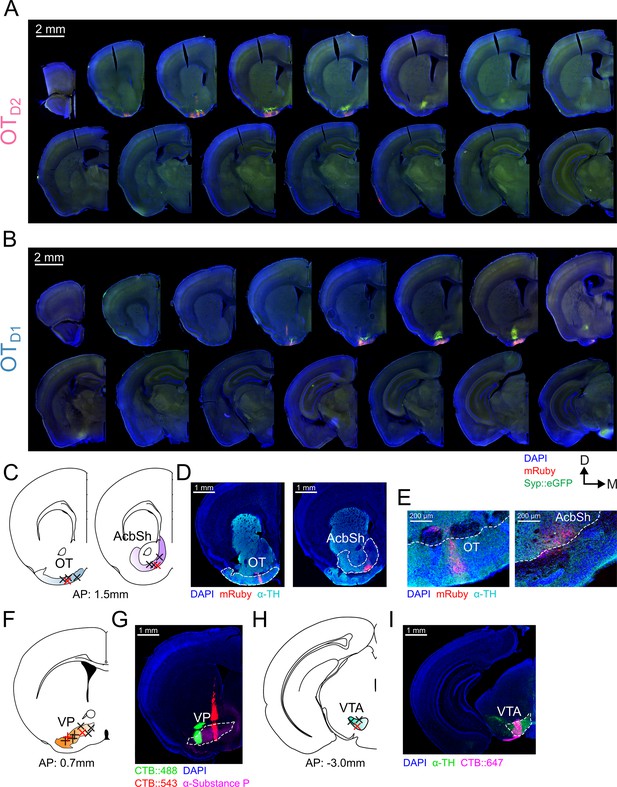
OTD1 and OTD2 primarily project to the lateral portion of the VP.
(A) Serial coronal sections from a representative experiment where AAVDJ-hSyn-FLEX-mRuby-T2A-syn-eGFP virus was injected into the anterior OT of an Adora2a-Cre mouse. Sections are roughly 400 µm apart from each other and range from +2.5mm to -3.0 mm relative to bregma. (B) Same as (A) but in a Drd1-Cre mouse. (C) Schematic showing the centroids of 4 injection sites for OT and AcbSh anterograde tracing experiments. (D, E) Representative images of the injection sites shown in (C). Sections were counterstained with ⍺-TH to delineate the boundary between the striatum and the rostral ventral pallidum. The centroids of these samples are marked as red x’s in (C). (F) Schematic showing the centroids of 4 CTB injection sites for lateral and medial VP. CTB::488 injection to the lateral VP are marked by +’s, whereas CTB::543 injection to the medial VP are marked by x’s. (G) Representative image of the injection sites shown in (F). Sections were counterstained with ⍺-Substance P to mark the boundary of the VP. The centroids of this sample are marked by the red +and red x in (F). (H) Schematic showing the centroids of 3 CTB injection sites for VTA. (I) Representative image of the injection sites shown in (H). Sections were counterstained with ⍺-TH to mark the boundary of the VTA.

Head-fixed two-photon Ca22+ imaging of OTD1, OTD2, or VP neurons during 6-odor conditioning paradigm.
(A) State-diagram of odor conditioning paradigm. Each trial begins with 2 s of odor delivery. Odors are chosen in pseudorandomized order such that the same odor is not repeated more than twice in a row. At the end of odor delivery, there is a variable delay (100–300ms), after which the animal is given either a 10% sucrose solution (SK and ST), a 70 psi airpuff (PK and PT), or nothing (XK and XT). Trials are separated by a variable intertrial interval (ITI; 12–18 s). Schematic representation of (B) lens implant surgery and (C) headfix two-photon microscopy setup. An example of spatial (D) and temporal (E) components extracted by CNMF from Drd1-Cre animal on day 3 of imaging. (D) The spatial footprints of 20 example neurons are shown on top of a maximum-correlation pixel image that was used to seed the factorization. The number displayed over each neuron matches the row number of the temporal components in (E). (F) An example raster plot (top) and averaged-across-trials trace (bottom) of the licking behavior recorded concurrently as (D) and (E). The timing of odor delivery is shown as shaded rectangles. The timing of US delivery is shown as arrowheads. (G) The mean total licks during each of the odors is shown averaged across all animals (n=17) after application of a moving-average filter with a window size of 10 trials. Red line marks the sucrose and airpuff contingency switch between day 3 and day 4. (H) Bar graph showing the licks during either sucrose cue expressed as a fraction of all licks during any odor. FWER-adjusted statistical significance for post hoc comparisons are shown as: ***p<0.001, **p<0.01, *p<0.05. See Appendix 1—tables 4 and 5 for detailed statistics.
Histological verification of lens implant location.
(A) Schematic showing the center, along the AP axis, of the implanted GRIN lens in six OTD2 jGCaMP7s animals. (B) Representative image of lens implant sites shown in (A). Sections were counterstained with ⍺-Substance P to delineate the boundary of the VP. This sample is marked by a red horizontal line in (A). (C) Schematic as in (A) for six OTD1 jGCaMP7s animals. (D) Representative image of lens implant sites shown in (C). (E) Schematic as in (A) for five VP jGCaMP7s animals. (F) Representative image of lens implant sites shown in (E). (G) Schematic as in (A) for 5 VP jGCaMP7s animals recorded during the lick spout retraction paradigm (Figure 6). (H) Representative image of lens implant sites shown in (G). Sections were counterstained with ⍺-Substance P to delineate the boundary of the VP. This sample is marked by a red horizontal line in (G).
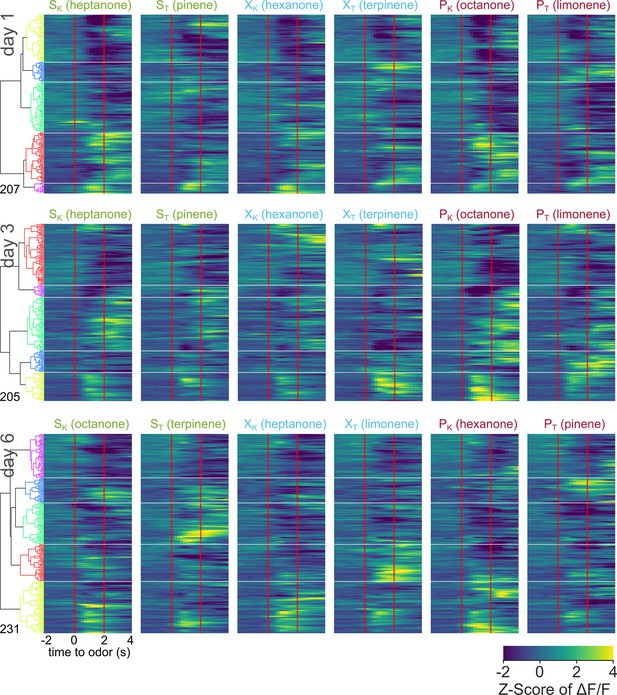
Pooled averaged-over-trials neural activity of all neurons from OTD2 animals across days.
Heatmap of odor-evoked activity in OTD2 neurons from day 1, day 3, and day 6 of imaging. The fluorescence measurements from each neuron were averaged over trials, Z-scored, then pooled for hierarchical clustering. Neurons are grouped by similarity, with the dendrogram shown on the right. Horizontal white lines demarcate the boundaries between the six clusters. Odor delivered at 0–2 s marked by vertical red lines. From left to right, the columns represent neural responses to sucrose-paired ketone and terpene, control ketone and terpene, and airpuff-paired ketone and terpene (SK, ST, XK, XT, PK, PT). Data is pooled from six animals.
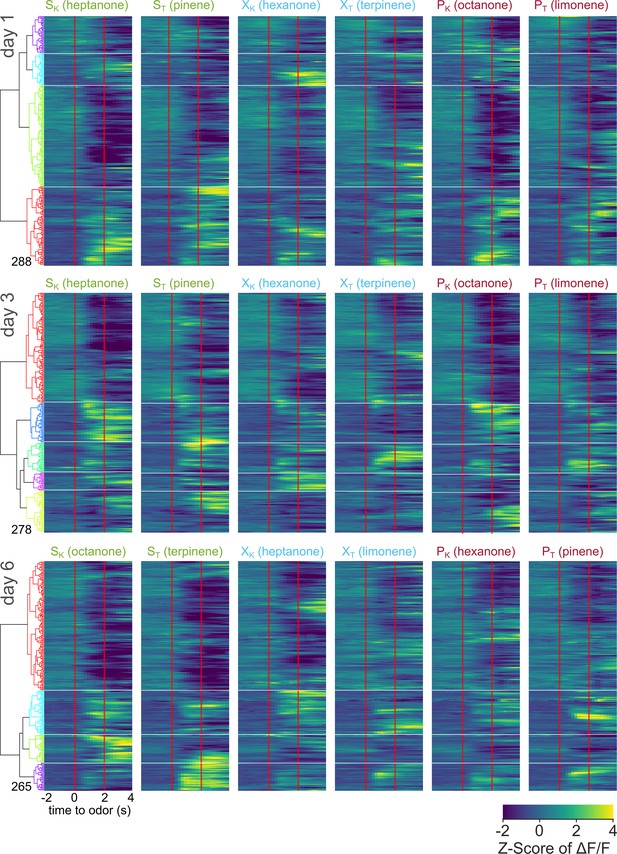
Pooled averaged-over-trials neural activity of all neurons from OTD1 animals across days.
Heatmap of odor-evoked activity in OTD1 neurons from day 1, day 3, and day 6 of imaging. The fluorescence measurements from each neuron were averaged over trials, Z-scored, then pooled for hierarchical clustering. Neurons are grouped by similarity, with the dendrogram shown on the right. Horizontal white lines demarcate the boundaries between the six clusters. Odor delivered at 0–2 s marked by vertical red lines. From left to right, the columns represent neural responses to sucrose-paired ketone and terpene, control ketone and terpene, and airpuff-paired ketone and terpene (SK, ST, XK, XT, PK, PT). Data is pooled from six animals.
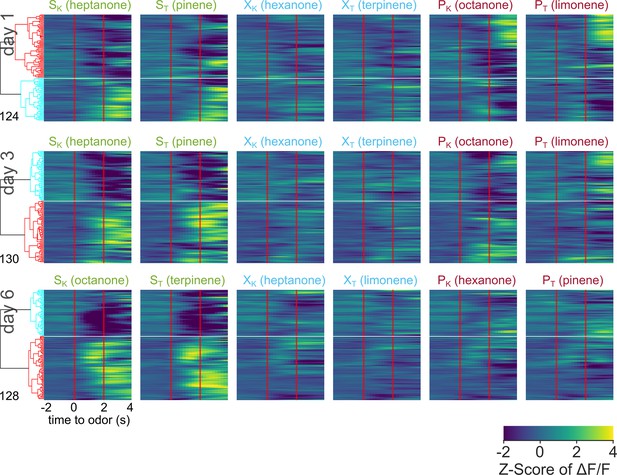
Pooled averaged-over-trials neural activity of all neurons from VP animals across days.
Heatmap of odor-evoked activity in VP neurons from day 1, day 3, and day 6 of imaging. The fluorescence measurements from each neuron were averaged over trials, Z-scored, then pooled for hierarchical clustering. Neurons are grouped by similarity, with the dendrogram shown on the right. Horizontal white lines demarcate the boundaries between the six clusters. Odor delivered at 0–2 s marked by vertical red lines. From left to right, the columns represent neural responses to sucrose-paired ketone and terpene, control ketone and terpene, and airpuff-paired ketone and terpene (SK, ST, XK, XT, PK, PT). Data is pooled from five animals.
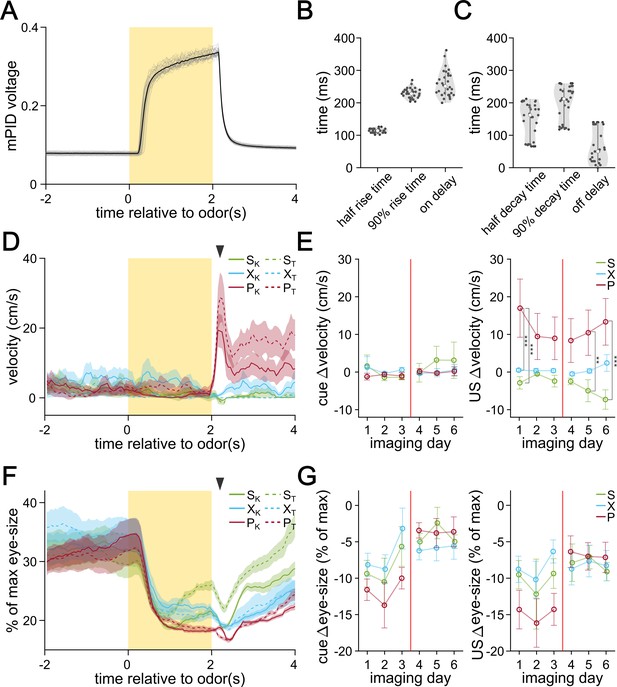
Extended behavioral analysis from imaging period.
(A) mPID voltage reading in response to 30 trials of a sample odor (α-terpinene) delivery. The time period during which the odor valve was turned on is shown by the yellow rectangle. Individual recordings are shown in gray and the average is shown in black. (B) On-kinetics of odor delivery. On delay refers to the interval between the valve turning on and the mPID voltage increasing by more than 10% of baseline. (C) Off-kinetics of odor delivery. Off delay refers to the interval between the odor valve turning off and the mPID voltage decreasing by more than 10% of its maximum. (D) Representative velocity of the head-fixed mouse in response to the six different odors measured by a digital encoder. The lines represent the average across 30 trials and the shaded areas represent the SEM. The black arrowhead marks when the US is delivered. (E) The difference in walking velocity in response to odor (left) and US delivery (right). Differences are calculated between the last second before odor delivery and the last second before the odor exposure (left) or the first half second after US delivery (right) grouped by US pairing. Circles represent the average across animals and the error bars show SEM. (F) Representative changes in range-normalized eye-size in response to the six different odors. (G) The difference in eye-size in response to odor (left) and US delivery (right). Differences are calculated between the last second before odor delivery and the last second before the odor exposure (left) or the first half second after US delivery (right) grouped by US pairing. Circles represent the average across animals and the error bars show SEM. FWER-adjusted statistical significance for post hoc comparisons are shown as: ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05. See Appendix 1—tables 32–36 for detailed statistics.
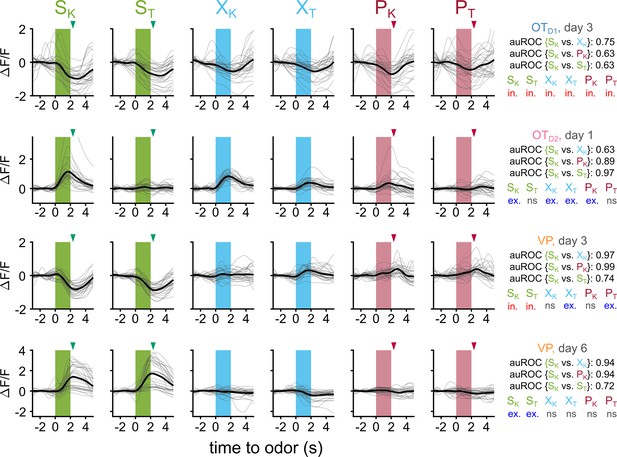
Traces of example neurons and their corresponding metrics.
(A) Example traces from an OTD1 neuron recorded on day 3. Each column shows this neuron’s response to a given odor across 30 trials (gray). The average across all trials is shown in black. For green and red arrowheads mark the time at which sucrose and airpuff were delivered, respectively. auROC values from single-neuron binary classifiers for discriminating {SK vs. XK}, {SK vs. PK}, and {SK vs. ST} are displayed on the right. Additionally, the results of statistical analysis to determine if this neuron reliably responded to each odor (in.=significant inhibitory response, exc.=significant excitatory response, ns = no significant difference between baseline and odor period). (B) Example traces from an OTD2 neuron recorded on day 1. (C) Example traces from a VP neuron recorded on day 3. (D) Example traces from a VP neuron recorded on day 6.
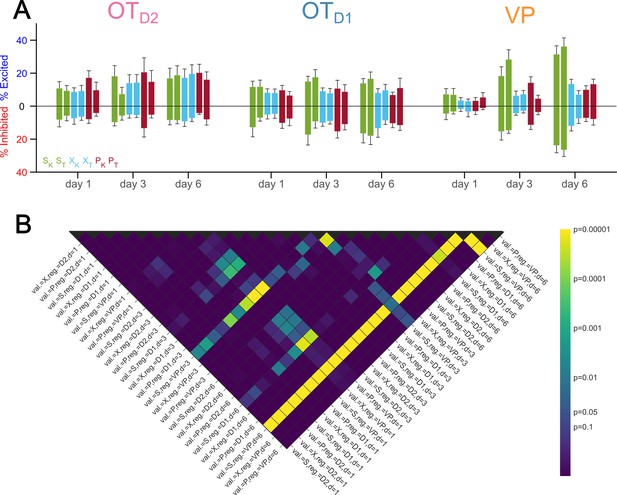
Percentage of neurons responsive to each odor across days.
(A) Bar graphs showing percentage of neurons from each region on imaging days 1, 3, and 6 that were significantly excited or inhibited by each odor. The average across animals is shown by the bar and the error bars represent SEM. (B) Heatmap of post hoc pairwise comparison p-values of percent responsive across imaging days and imaging region. See Appendix 1—table 37 for detailed statistics.
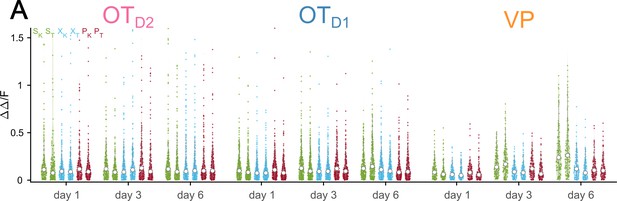
Distribution of response magnitudes to each odor across days.
Violin plots showing the averaged-over-trials response magnitudes to each odor during the last second of odor exposure. See Appendix 1—table 38 for detailed statistics.

VP neurons encode reward-contingency more robustly than OTD1 or OTD2 neurons.
(A) Heatmap of odor-evoked activities in OTD1, OTD2, and VP neurons from day 6 of imaging. The fluorescence measurements from each neuron were averaged over trials, Z-scored, then pooled for hierarchical clustering. Neurons are grouped by similarity, with the dendrogram shown on the right and a raster plot on the left indicating which region a given neuron is from. Horizontal white lines demarcate the boundaries between the 6 clusters. Odor delivered at 0–2 s marked by vertical red lines and US delivery is marked by arrowheads. From left to right, the columns represent neural responses to sucrose-paired ketone and terpene, control ketone and terpene, and airpuff-paired ketone and terpene (SK, ST, XK, XT, PK, PT). (B) Average Z-scored activity of each cluster to each of the six odors on day 6 of imaging. Yellow bar indicates 2 s of odor exposure. (C) The distribution of clusters by population. (D) Percentage of total neurons that were significantly excited or inhibited by each odor (Bonferroni-adjusted FDR <0.05) as a function of time relative to odor. Lines represent the mean across biological replicates and the shaded area reflects the mean ± SEM. (E) Bar graph showing % of neurons from each population that are responsive to both sucrose-paired odors in the same direction (left), responsive to only a single odor (middle), or responsive to at least 3 odors (right). Bars represent the mean across biological replicates and x’s mark individual animals. (F) Scatterplot comparing the magnitudes of SK responses (∆∆SK) to ST responses (∆∆ST). The dotted line represents the hypothetical scenario where ∆∆SK = ∆∆ST. For each population, the R2 value of the 2-d distribution compared to the ∆∆SK = ∆∆ST line is reported. (G) Same as F but comparing ∆∆SK to ∆∆XK. (H) Lineplot showing the % of neurons from each population where the difference between ∆∆SK and ∆∆XK is lower than that between ∆∆SK and ∆∆ST. (I) Bargraph showing % of neurons whose responses to {SK vs. XK} can be discriminated by a linear classifier with auROC >0.75. (J) Same as (I) but for {SK vs PK}. (K) Same as (I) but for {SK vs ST}. (L) Schematic representation of four possible categories for a joint-distribution of {SK vs. XK} and {SK vs. ST} auROC values. Identity-encoding neurons could be in any quadrant other than the bottom-left, whereas valence-encoding neurons should be in the bottom-right quadrant. (M) Scatterplot of each neuron’s auROC value for {SK vs. XK} on the x-axis and {SK vs. ST} on the y-axis on days 1, 3, and 6 of imaging. (N) Stacked bar graph showing the distribution of neurons from each population that fall into each of the four quadrants across the 3 different imaging days. FWER-adjusted statistical significance for post hoc comparisons are shown as: ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05. See Appendix 1—tables 6–17 for detailed statistics. Source data available at 10.5061/dryad.2547d7x15.
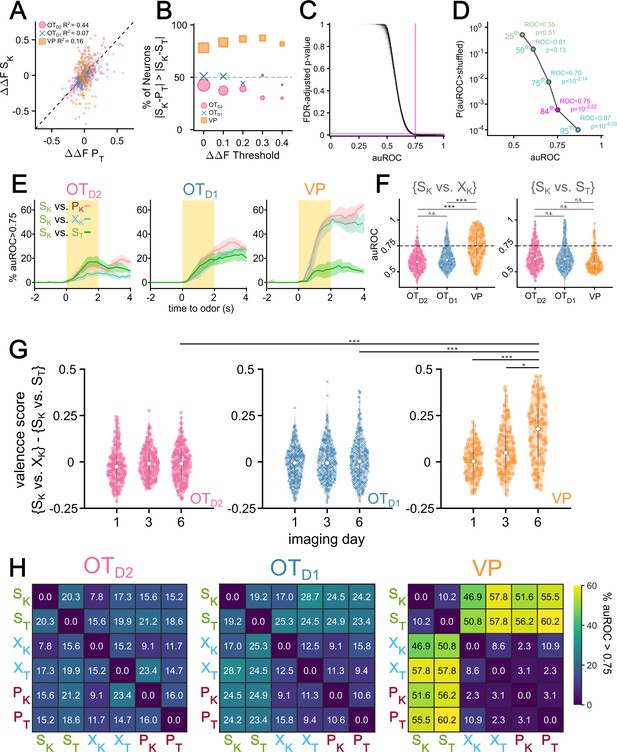
Pairwise analysis of single neuron odor encoding.
(A) Scatterplot comparing the magnitudes of SK responses (∆∆SK) to PT responses (∆∆PT). The dotted line represents the hypothetical scenario where ∆∆SK = ∆∆PT. For each population, the R2 value of the 2-d distribution compared to the ∆∆SK = ∆∆PT line is reported. (B) The percentage of neurons from each population where the difference between ∆∆SK and ∆∆PT is lower than that between ∆∆SK and ∆∆ST. (C) Bootstrapped FDR-adjusted p-values as a function of auROC values of single-neuron binary classifiers. In total, there are 27,900 single-neuron binary classifiers (15 pairwise classifiers for each of the 1860 recordings across three regions and days 1, 3, and 6 of imaging). Each classifier was compared against 10,000 shuffles. Horizontal magenta line marks FDR-adjusted p-value of 0.001 and the vertical magenta line marks auROC of 0.75. (D) The 25th, 50th, 75th, 84th, and 95th percentiles of auROC values and their corresponding unadjusted p-values. For auROC values that were greater than all 10,000 shuffles, a conservative p-value of 0.0001 was assigned. (E) The percentage of day 6 {SK vs PK}, {SK vs XK}, and {SK vs ST} auROC values greater than 0.75 as a function of time relative to odor, grouped by region. Lines represent the average across biological replicates and the shaded area shows the SEM. (F) Violin plot showing the distribution of pooled day 6 {SK vs XK} (left) and {SK vs ST} (right) auROC values grouped by region. Horizontal dotted line marks auROC = 0.75. (G) Violin plot of the distribution of single-neuron valence scores (defined as the difference between the average auROC for {S vs. X|P} classification and {SK vs. ST} classification), grouped by imaging day and region. (H) Heatmap of percentage of single-neuron pairwise classifiers with auROC >0.75. Classifiers were trained from neural activity recorded during the last second of odor exposure. Percentage of neurons with auROC >0.75 for a given binary classification was averaged across animals and grouped by region. For post hoc pairwise comparisons, the median values for all neurons in each animal were compared across imaging day and region. The FWER-adjusted p-values are shown as: ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05. See Appendix 1—tables 39–46 for detailed statistics.
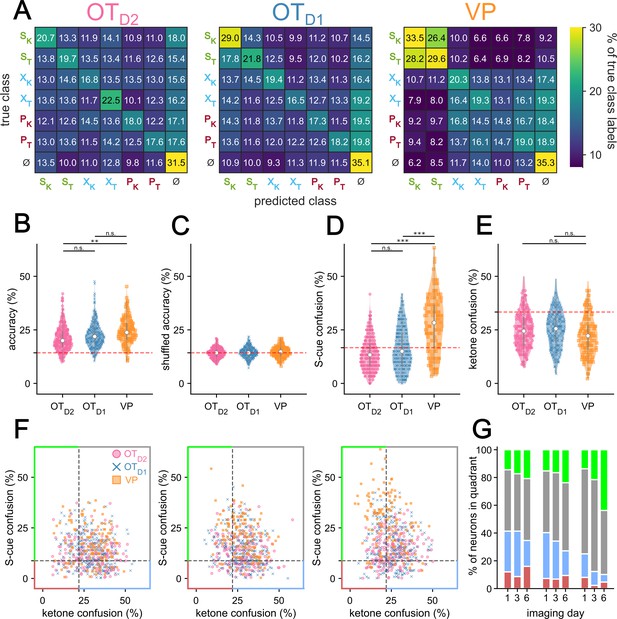
Multinomial analysis of single neuron odor encoding.
(A) Confusion matrix of single-neuron MNR classifiers trained on neural activity during the last second of odor exposure on day 6 of imaging. Rows represent the true class while columns represent the predicted class. Each confusion matrix is averaged across 10 k-fold and across all neurons of a given region. ɸ represents data taken from 30 pre-odor bins randomly sampled from –1.5 to –0.5 s relative to odor delivery. (B) Violin plot of single-neuron MNR classifier accuracy, averaged across 10 k-fold, grouped by region. (C) Violin plot of the single-neuron MNR classifier accuracy trained on shuffled data. Each data point represents the average across 10 shuffles. (D) Violin plot of MNR S-cue confusion, that is confusion between SK and ST. This corresponds to (1) when the true class was SK but predicted class was ST and (2) when the true class was ST but the predicted class was SK. (E) Violin plot of MNR confusion among all ketones. This corresponds when the true class was a ketone and the predicted class was a different ketone (e.g. true class = XK and predicted class = PK). (F) Scatterplot of each neuron’s ketone confusion on the x-axis and S-cue confusion on the y-axis on days 1, 3, and 6 of imaging. (G) Stacked bar graph showing the distribution of neurons from each population that fall into each of the four quadrants across the 3 different imaging days. For post hoc pairwise comparisons, the median values for all neurons in each animal were compared across imaging day and region. The FWER-adjusted p-values are shown as: ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05. See Appendix 1—tables 47–53 for detailed statistics.
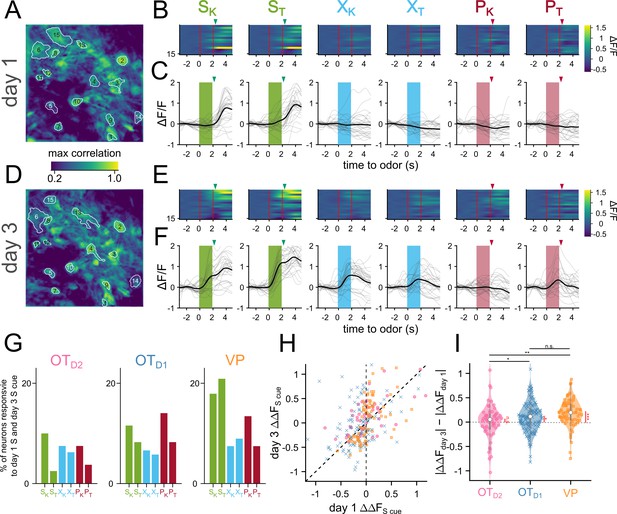
Sucrose responsive VP neurons become sucrose-cue responsive after pairing.
(A) The spatial footprints of 15 neurons from day 1 are outlined over a max-correlation projection image. (B) Heatmap of averaged-over-trials ΔF/F in response to 6 odors on day 1. Odor delivery period is shown with 2 red vertical lines and sucrose/airpuff timing is shown with downward arrowhead. (C) An example neuron’s responses on day 1 across 30 trials to 6 different odors. Individual trial traces are shown in light gray whereas the averaged-across trials trace is shown in black. Odor delivery period is depicted as shaded rectangles and US delivery is marked by arrowheads. (D–F) Same as (A–C), respectively, but for day 3. (G) Percentage of all tracked neurons that were both sucrose-responsive on day 1 and odor-responsive in the same direction on day 3. (H) Scatter plot of averaged-over-trials responses to SK or ST on day 1 (x-axis) and day 3 (y-axis). Each point is a neuron that was successfully matched from day 1 and day 3. Neurons from OTD2, OTD1, and VP are plotted as pink circles, blue crosses, and yellow squares, respectively. Neurons that have increased response magnitudes on day 3 would fall between the two dotted lines. (I) Violin plot showing the distributions of day 3 responsive magnitude – day 1 response magnitude. Black asterisks show statistical significance of pairwise comparisons and red asterisks show statistical significance for one-sample t-tests. Pairwise comparisons were done using the Student’s t-test. The p-values were corrected for FDR by Benjamini-Hocherg procedure. ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05. See Appendix 1—tables 18 and 19 for detailed statistics. Source data available at 10.5061/dryad.2547d7x15.

OT encodes odor identity in high-dimensional space and VP encodes reward-contingency in low-dimensional space.
(A) Average normalized pairwise Euclidean distance between odor-evoked population-level activity from day 6 of imaging shown as a function of time relative to odor delivery. Traces show the average value across biological replicates of the same population and the shaded areas represent the average ± SEM. (B) A heatmap of the average normalized pairwise distance during the odor delivery period. (C) Average CV accuracy of binary pairwise linear classifiers trained on population data plotted against time relative to odor delivery. (D). A heatmap of the average CV accuracy during the odor delivery period. (E) Schematic representation of generalized linear classification performance for an idealized valence encoder. Each row corresponds to the training odor-pair and each column corresponds to the testing odor-pair. For an idealized valence encoder, the decodability would generalize well across odor-pairs of the equal valence grouping outlined in red. Note that the elements along the diagonal are cases where training and testing odor-pairs are identical and do not reflect generalizability. (F) Heatmap representing the maximum generalized linear classification accuracy during odor delivery period averaged across biological replicates for each population. (G) Mean cross-validated linear classifier accuracy for S-cue vs. control or puff-cue classification and the generalized accuracy for S-cue vs. control or puff-cue classification after training on a different pair. Bar represents the mean across biological replicates and x’s mark accuracy values for individual animals. (H) Average PR normalized to n calculated after randomly subsampling an increasing number of neurons. (I) Average PR calculated after subsampling 15 neurons. (J) Average CV accuracy of linear classifiers trained on {SK vs. PK} plotted against number of principal components used for training. For each simultaneously imaged group of neurons, 15 neurons were subsampled and classifiers were trained on an increasing number of principal components. Thinner faded lines show mean accuracy across subsampling for individual animals. Markers represent the mean across biological replicates. Error bars indicate SEM across biological replicates. (K) Average CV accuracy of linear classifiers trained on {SK vs. ST}. (L) Comparison of the average accuracy of {SK vs. PK} classifiers trained on the 1st PC vs. {SK vs. ST} classifiers trained on all 15 PCs. FWER-adjusted statistical significance for post hoc comparisons are shown as: ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05. See Appendix 1—tables 20–29 for detailed statistics. Source data available at 10.5061/dryad.2547d7x15.
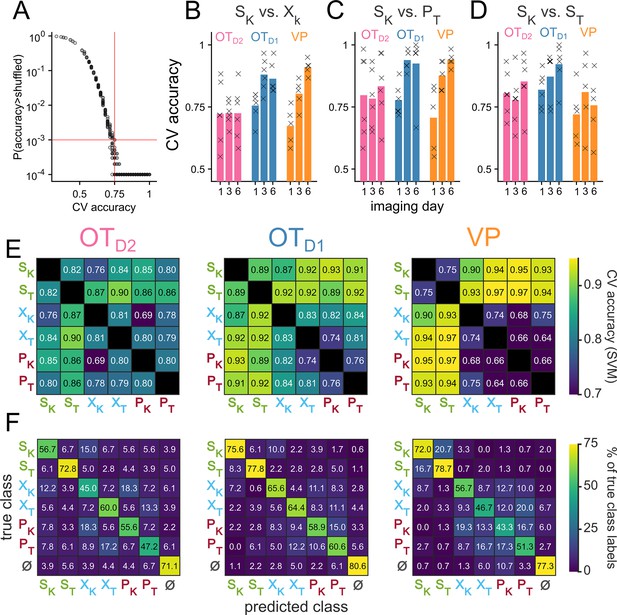
Analysis of population-level odor encoding.
(A) Scatterplot of CV-accuracy of linear classifiers trained on simultaneously-recorded neurons on the x-axis and their bootstrapped unadjusted p-values on the y-axis. Red horizontal line marks p=0.001 and red red vertical line marks CV accuracy = 0.75. All classifiers with CV accuracy higher than 0.75 had p<0.001. In total, there are 765 binary classifiers (15 pairwise classifiers for each of the 51 recordings across 3 regions and days 1, 3, and 6 of imaging). Each classifier was compared against 10,000 shuffles. For auROC values that were greater than all 10,000 shuffles, a conservative p-value of 0.0001 was assigned. (B) The CV accuracy for {SK vs XK} binary classification trained on the last second of population-level activity. The bars represent the average across biological replicates. CV accuracy from individual animals are shown as x’s. (C) Same as (B) but for {SK vs. PT} classification. (D) Same as (B) but for {SK vs. ST} classification. (E) Heatmap of CV accuracy from binary SVM’s trained on day 6 of imaging with a radial basis function kernel. CV accuracy was averaged across biological replicates. (F) Confusion matrix of population-level MNR classifiers trained on neural activity during the last second of odor exposure on day 6 of imaging. Rows represent the true class while columns represent the predicted class. Each confusion matrix is averaged across biological replicates. ɸ represents data taken from 30 pre-odor bins randomly sampled from –1.5 to –0.5 seconds relative to odor delivery. See Appendix 1—tables 54–59 for details on statistical comparison of average classifier accuracy across animals.

Separate VP populations encode reward-contingency and licking vigor.
(A) State diagram for odor pairing paradigm where lick spout is removed during the presentation of half of the odors. The paradigm is similar to one described in Figure 2A with the following key differences: (1) the lick spout is moved away from the animal’s mouth during the presentation of half of the odors (Nhi, Nlo, NX). (2) sucrose is delivered after a longer variable delay (1.1–1.3 s). (3) 2 of the odors have 100% sucrose contingency (Lhi, Nhi), 2 of the odors have 50% sucrose contingency (Llo, Nlo), and the other 2 have 0% sucrose contingency (LX, NX). (B) Schematic showing the timing of lick port movement relative to odor and sucrose delivery. (C) Licking behavior to 6 odors averaged across 30 trials from a representative animal. Duration of odor delivery is marked by the shaded rectangle and the average time of sucrose delivery is marked by the arrowhead. The time bin used for subsequent analysis (last 0.5 s of odor and first 0.5 s of delay) is outlined by square brackets (D) Average licks/s for each odor measured between the last 0.5 s of odor and the first 0.5 s of delay. Data were pooled from the day of highest difference between licks to Lhi and Nhi. (E) Heatmap of odor-evoked activity in VP neurons pooled from each animal’s day of highest difference between licks to Lhi and Nhi. Neurons are grouped according to the clustering dendrogram, shown on the right. Horizontal white lines demarcate the boundaries between the three clusters. Odor delivery is marked by vertical red lines. (F) Average Z-scored activity of each cluster to each of the six odors. Yellow bar indicates 2 s of odor exposure. (G) The percentage of single-neuron linear classifiers with auROC >0.75 as a function of time relative to odor delivery. Shaded area represents the SEM across biological replicates (n=5). (H) Heatmap of the percentage of pooled VP neurons with auROC >0.75 during the last 0.5 s of odor and first 0.5 s of delay. (I) Scatterplot comparing the auROC for {Lhi vs Nhi} (y-axis) and {Nhi vs. NX} (x-axis) for each neuron. The line of best fit is plotted as a dotted line, with the 95% confidence interval shaded in. (J) Same as (I) but comparing the auROC for {Lhi vs LX} (y-axis) and {Nhi vs. NX} (x-axis). (K) Scatterplot comparing regression models that explain each neuron’s activity on a given trial as a function of anticipatory licking or sucrose contingency. The values plotted are the loss in R2 in models without anticipatory licking (y-axis) or sucrose contingency (x-axis) when compared to a model with both variables and their interaction term. (L) CV accuracy for five different odor pairs as a function of time relative to odor delivery. (M) Heatmap of average pairwise CV accuracy trained on the last 0.5 s of odor and the first 0.5 s of delay. (N) Scatterplot of all pairwise classifier accuracies from all animals (y-axis) and the corresponding range-normalized average pairwise difference in anticipatory licking (x-axis). (O) Scatterplot of all pairwise classifier accuracies from all animals (y-axis) and the corresponding pairwise difference in reward-contingency (x-axis). (P) Scatterplot of all pairwise classifier accuracies (y-axis) and the adjusted combined model of ranged-normalized Δlick and Δreward-contingency (x-axis). FWER-adjusted statistical significance for post hoc comparisons are shown as: ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05. See Appendix 1—tables 30 and 31 for detailed statistics. Source data available at10.5061/dryad.2547d7x15.
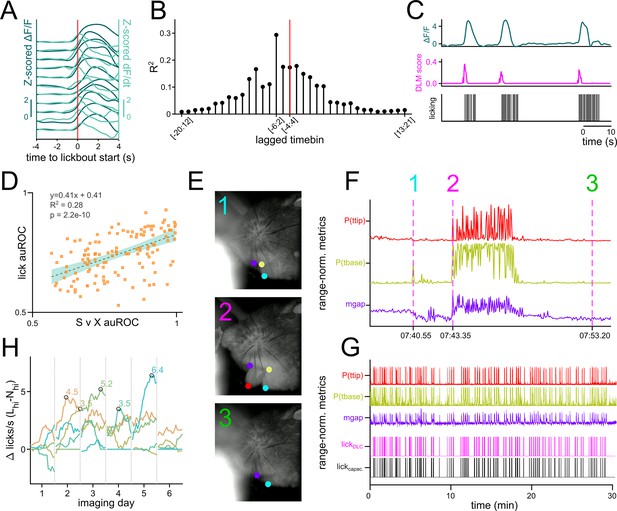
Camera-based detection of licking in head-fixed animals.
(A–C) Metrics of a representative neurons with activity that predicts licking. (A) Representative neuron’s Z-scored ΔF/F (dark green) and Z-scored dF/dt (light green) aligned to the onset of a lickbout. Each line represents the same neuron’s activity during an individual lickbout. (B) Stemplot showing an example of the lagged correlation between the onset of licking and the fluorescence of a neuron across 9 frames (1 frame = 0.2 s). Time bins of various lags are shown on the x-axis (negative number denotes frames that precede onset of licking) and the resulting R2 is plotted on the y-axis. Red vertical line marks the case where the 9 frames are centered on the onset of licking. As an example, [–6:2] refers to fluorescence between 1.2 s prior to lickbout onset and 0.4 seconds after lickbout onset. (C) The output of a distributed lag model (DLM) that predicts the onset of a lickbout from ΔF/F of a single neuron. ΔF/F (dark green, top), DLM score (magenta, middle), and the licking recorded by a capacitive sensor (black, bottom) are shown in parallel. The DLM model was trained using 9 distributed frames ([–6:2]) of ΔF/F for each frame of lickbout onset. (D) Scatterplot of day 6 VP neurons’ DLM lick classifier auROC on the y-axis plotted against their mean {SK vs. XK} auROC on the x-axis. The slope, intercept, R2, and p-value of the slope are shown on the top left corner. (E) Three example snapshots of the camera feed during moving lick spout paradigm with overlay of DeepLabCut labeling. The coordinates of top lip, bottom lip, base of tongue (tbase), and tip of tongue (ttip) are displayed with a probability cutoff of 0.4. (F) Range-normalized metrics from DLC labeling. P(ttip) (red, top) is the probability score assigned to the labeling of the tongue tip. P(tbase) (yellow, middle) is the probability score assigned to the labeling of the tongue base. Mgap (purple, bottom) is the Euclidean distance between the top lip and the bottom lip. The three vertical magenta lines represent the timing of the three snapshots shown in (E). (G) The same range-normalized metrics as in (F) plotted against the ground truth licking data from capacitive sensor (black, bottom) and DLC-based licking classifier score (magenta, second from bottom). (H) Lineplot showing the difference in total licking to Lhi and Nhi during the time bin (1.5–2.5 s after odor onset) used for most analyses plotted against imaging day for individual animals. The time of peak difference is circled in black.
Tables
Pairwise comparisons of anterograde labeling from OT and AcbSh (Figure 1C).
| Group A | Group B | Lower Limit | A-B | Upper Limit | FDR adjusted p-value |
|---|---|---|---|---|---|
| OTD1 → VP | AcbD1 → VP | –20.506 | –8.798 | 2.910 | 0.577 |
| OTD1 → VP | OTD2 → VP | –0.851 | 9.922 | 20.694 | 0.577 |
| OTD1 → VP | AcbD2 → VP | 9.338 | 19.340 | 29.341 | 0.291 |
| AcbD1 → VP | OTD2 → VP | 11.163 | 18.719 | 26.275 | 0.161 |
| AcbD1 → VP | AcbD2 → VP | 21.729 | 28.137 | 34.545 | 3.394E-02 |
| OTD2 → VP | AcbD2 → VP | 4.942 | 9.418 | 13.894 | 0.206 |
| OTD1 → VP | AcbD1 → VP | –9.944 | –8.199 | –6.453 | 2.223E-02 |
| OTD1 → LH | OTD2 → LH | –0.045 | 0.564 | 1.173 | 0.577 |
| OTD1 → LH | AcbD2 → LH | –0.016 | 0.591 | 1.198 | 0.577 |
| AcbD1 → LH | OTD2 → LH | 7.125 | 8.763 | 10.401 | 2.223E-02 |
| AcbD1 → LH | AcbD2 → LH | 7.153 | 8.790 | 10.426 | 2.223E-02 |
| OTD2 → LH | AcbD2 → LH | –0.029 | 0.027 | 0.082 | 0.655 |
| OTD1 → VTA | AcbD1 → VTA | –17.357 | –14.504 | –11.650 | 2.223E-02 |
| OTD1 → VTA | OTD2 → VTA | –0.022 | 0.183 | 0.387 | 0.577 |
Pairwise comparisons of retrograde labeling from vlVP and dmVP (Figure 1F).
| Group A | Group B | Lower Limit | A-B | Upper Limit | FDR adjusted p-value |
|---|---|---|---|---|---|
| AI→vlVP | AI→dmVP | 78.249 | 148.500 | 218.751 | 0.116 |
| Acb→vlVP | Acb→dmVP | –185.929 | –123.250 | –60.571 | 0.116 |
| LS→vlVP | LS→dmVP | –130.496 | –118.750 | –107.004 | 3.27E-04 |
| OFC→vlVP | OFC→dmVP | –167.021 | –132.000 | –96.979 | 1.86E-02 |
| OT→vlVP | OT→dmVP | 179.793 | 221.750 | 263.707 | 5.57E-03 |
| Pir→vlVP | Pir→dmVP | –71.946 | –21.500 | 28.946 | 0.685 |
Pairwise comparisons of retrograde labeling from VTA (Figure 1I).
| Group A | Group B | Lower Limit | A-B | Upper Limit | FDR adjusted p-value |
|---|---|---|---|---|---|
| OT→VTA | AcbSh→VTA | –735.101 | –575 | –414.899 | 3.44E-02 |
| OT→VTA | AcbC→VTA | –1027.381 | –915 | –802.619 | 3.71E-03 |
| AcbC→VTA | AcbSh→VTA | 144.452 | 340 | 535.548 | 0.157 |
Two-way ANOVA for effect of day or lens placement on licking accuracy (Figure 2H).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| day | 4.301 | 5 | 0.860 | 27.638 | 2.29E-16 |
| region | 0.143 | 2 | 0.072 | 2.301 | 0.106 |
| day:region | 0.251 | 10 | 0.025 | 0.806 | 0.623 |
| Error | 2.583 | 83 | 0.031 | ||
| Total | 7.305 | 100 |
Post hoc pairwise comparisons of licking accuracy across imaging days (Figure 2H).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| day 1 | day 2 | –0.385 | –0.208 | –0.031 | 1.18E-02 |
| day 1 | day 3 | –0.632 | –0.452 | –0.272 | 2.03E-09 |
| day 1 | day 4 | –0.134 | 0.043 | 0.220 | 0.980 |
| day 1 | day 5 | –0.558 | –0.381 | –0.203 | 2.30E-07 |
| day 1 | day 6 | –0.650 | –0.473 | –0.295 | 2.58E-10 |
| day 2 | day 3 | –0.424 | –0.244 | –0.064 | 2.18E-03 |
| day 2 | day 4 | 0.074 | 0.251 | 0.429 | 1.14E-03 |
| day 2 | day 5 | –0.350 | –0.172 | 0.005 | 0.061 |
| day 2 | day 6 | –0.441 | –0.264 | –0.087 | 5.33E-04 |
| day 3 | day 4 | 0.315 | 0.495 | 0.675 | 8.31E-11 |
| day 3 | day 5 | –0.109 | 0.071 | 0.251 | 0.856 |
| day 3 | day 6 | –0.200 | –0.021 | 0.159 | 0.999 |
| day 4 | day 5 | –0.601 | –0.424 | –0.247 | 1.00E-08 |
| day 4 | day 6 | –0.693 | –0.516 | –0.338 | 9.77E-12 |
| day 5 | day 6 | –0.269 | –0.092 | 0.085 | 0.657 |
2-way ANOVA for effect of day or lens placement on percentage of neurons responsive to a single odor (Figure 3E).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| day | 0.05917 | 2 | 0.02958 | 2.99563 | 0.06079 |
| region | 0.03696 | 2 | 0.01848 | 1.87142 | 0.16651 |
| day:region | 0.06481 | 4 | 0.01620 | 1.64062 | 0.18197 |
| Error | 0.41479 | 42 | 0.00988 | ||
| Total | 0.57170 | 50 |
Post hoc pairwise comparisons of percentage of neurons responsive to a single odor across imaging days and region (Figure 3E).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,D2 OT | d3,D2 OT | –0.140 | 0.048 | 0.236 | 0.995 |
| d1,D2 OT | d6,D2 OT | –0.164 | 0.024 | 0.211 | 1.000 |
| d3,D2 OT | d6,D2 OT | –0.212 | –0.024 | 0.163 | 1.000 |
| d1,D1 OT | d3,D1 OT | –0.287 | –0.099 | 0.088 | 0.724 |
| d1,D1 OT | d6,D1 OT | –0.328 | –0.141 | 0.047 | 0.285 |
| d3,D1 OT | d6,D1 OT | –0.229 | –0.041 | 0.146 | 0.998 |
| d1,VP | d3,VP | –0.321 | –0.115 | 0.090 | 0.662 |
| d1,VP | d6,VP | –0.335 | –0.129 | 0.076 | 0.513 |
| d3,VP | d6,VP | –0.220 | –0.014 | 0.191 | 1.000 |
| d1,D2 OT | d1,D1 OT | –0.115 | 0.072 | 0.260 | 0.937 |
| d1,D2 OT | d1,VP | –0.056 | 0.141 | 0.338 | 0.341 |
| d1,D1 OT | d1,VP | –0.128 | 0.069 | 0.265 | 0.964 |
| d3,D2 OT | d3,D1 OT | –0.263 | –0.075 | 0.112 | 0.923 |
| d3,D2 OT | d3,VP | –0.219 | –0.022 | 0.175 | 1.000 |
| d3,D1 OT | d3,VP | –0.144 | 0.053 | 0.250 | 0.993 |
| d6,D2 OT | d6,D1 OT | –0.280 | –0.092 | 0.095 | 0.796 |
| d6,D2 OT | d6,VP | –0.209 | –0.012 | 0.184 | 1.000 |
| d6,D1 OT | d6,VP | –0.117 | 0.080 | 0.276 | 0.918 |
Two-way ANOVA for effect of day or lens placement on percentage of neurons responsive to three or more odors (Figure 3E).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| day | 0.10897 | 2 | 0.05448 | 4.58607 | 0.01580 |
| region | 0.00469 | 2 | 0.00234 | 0.19732 | 0.82168 |
| day:region | 0.06640 | 4 | 0.01660 | 1.39730 | 0.25144 |
| Error | 0.49897 | 42 | 0.01188 | ||
| Total | 0.66598 | 50 |
Post hoc pairwise comparisons of percentage of neurons responsive to three or more odors across imaging days and region (Figure 3E).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,D2 OT | d3,D2 OT | –0.237 | –0.031 | 0.175 | 1.000 |
| d1,D2 OT | d6,D2 OT | –0.246 | –0.040 | 0.165 | 0.999 |
| d3,D2 OT | d6,D2 OT | –0.215 | –0.009 | 0.196 | 1.000 |
| d1,D1 OT | d3,D1 OT | –0.230 | –0.024 | 0.182 | 1.000 |
| d1,D1 OT | d6,D1 OT | –0.269 | –0.063 | 0.143 | 0.984 |
| d3,D1 OT | d6,D1 OT | –0.245 | –0.039 | 0.167 | 0.999 |
| d1,VP | d3,VP | –0.406 | –0.181 | 0.044 | 0.207 |
| d1,VP | d6,VP | –0.453 | –0.228 | –0.003 | 0.046 |
| d3,VP | d6,VP | –0.272 | –0.047 | 0.178 | 0.999 |
| d1,D2 OT | d1,D1 OT | –0.220 | –0.014 | 0.191 | 1.000 |
| d1,D2 OT | d1,VP | –0.124 | 0.092 | 0.308 | 0.894 |
| d1,D1 OT | d1,VP | –0.109 | 0.106 | 0.322 | 0.793 |
| d3,D2 OT | d3,D1 OT | –0.213 | –0.007 | 0.198 | 1.000 |
| d3,D2 OT | d3,VP | –0.274 | –0.058 | 0.158 | 0.993 |
| d3,D1 OT | d3,VP | –0.266 | –0.051 | 0.165 | 0.997 |
| d6,D2 OT | d6,D1 OT | –0.243 | –0.037 | 0.168 | 1.000 |
| d6,D2 OT | d6,VP | –0.311 | –0.096 | 0.120 | 0.871 |
| d6,D1 OT | d6,VP | –0.274 | –0.059 | 0.157 | 0.993 |
Two-way ANOVA for effect of day or lens placement on percentage of neurons responsive to both S-cues (Figure 3E).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| day | 0.11460 | 2 | 0.05730 | 6.60475 | 0.00321 |
| region | 0.18370 | 2 | 0.09185 | 10.58706 | 0.00019 |
| day:region | 0.16522 | 4 | 0.04131 | 4.76096 | 0.00294 |
| Error | 0.36438 | 42 | 0.00868 | ||
| Total | 0.80767 | 50 |
Post hoc pairwise comparisons of percentage of neurons responsive to both S-cues across imaging days and region (Figure 3E).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,D2 OT | d3,D2 OT | –0.139 | 0.037 | 0.213 | 0.999 |
| d1,D2 OT | d6,D2 OT | –0.200 | –0.024 | 0.151 | 1.000 |
| d3,D2 OT | d6,D2 OT | –0.237 | –0.062 | 0.114 | 0.963 |
| d1,D1 OT | d3,D1 OT | –0.239 | –0.063 | 0.112 | 0.957 |
| d1,D1 OT | d6,D1 OT | –0.196 | –0.020 | 0.156 | 1.000 |
| d3,D1 OT | d6,D1 OT | –0.133 | 0.043 | 0.219 | 0.996 |
| d1,VP | d3,VP | –0.441 | –0.248 | –0.056 | 3.79E-03 |
| d1,VP | d6,VP | –0.473 | –0.281 | –0.088 | 7.12E-04 |
| d3,VP | d6,VP | –0.225 | –0.032 | 0.160 | 1.000 |
| d1,D2 OT | d1,D1 OT | –0.189 | –0.013 | 0.163 | 1.000 |
| d1,D2 OT | d1,VP | –0.151 | 0.033 | 0.217 | 1.000 |
| d1,D1 OT | d1,VP | –0.138 | 0.046 | 0.230 | 0.996 |
| d3,D2 OT | d3,D1 OT | –0.289 | –0.114 | 0.062 | 0.480 |
| d3,D2 OT | d3,VP | –0.437 | –0.252 | –0.068 | 1.72E-03 |
| d3,D1 OT | d3,VP | –0.323 | –0.139 | 0.045 | 0.279 |
| d6,D2 OT | d6,D1 OT | –0.185 | –0.009 | 0.167 | 1.000 |
| d6,D2 OT | d6,VP | –0.408 | –0.223 | –0.039 | 7.94E-03 |
| d6,D1 OT | d6,VP | –0.399 | –0.214 | –0.030 | 1.23E-02 |
Two-way ANOVA for effect of day or lens placement on percentage of neurons with auROC >0.75 for {SK vs. PK} (Figure 3I).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| day | 0.85429 | 2 | 0.42714 | 46.23443 | 2.439E-11 |
| region | 0.32224 | 2 | 0.16112 | 17.43954 | 3.064E-06 |
| day:region | 0.40111 | 4 | 0.10028 | 10.85411 | 3.918E-06 |
| Error | 0.38802 | 42 | 0.00924 | ||
| Total | 1.87808 | 50 |
Post hoc comparisons of percentage of neurons with auROC >0.75 for {SK vs. PK} across imaging day and region (Figure 3I).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,D2 OT | d3,D2 OT | –0.259 | –0.078 | 0.103 | 0.890 |
| d1,D2 OT | d6,D2 OT | –0.279 | –0.097 | 0.084 | 0.709 |
| d3,D2 OT | d6,D2 OT | –0.201 | –0.020 | 0.162 | 1.000 |
| d1,D1 OT | d3,D1 OT | –0.293 | –0.112 | 0.069 | 0.541 |
| d1,D1 OT | d6,D1 OT | –0.417 | –0.236 | –0.054 | 0.003 |
| d3,D1 OT | d6,D1 OT | –0.305 | –0.124 | 0.058 | 0.407 |
| d1,VP | d3,VP | –0.476 | –0.278 | –0.079 | 0.001 |
| d1,VP | d6,VP | –0.820 | –0.621 | –0.423 | 2.00E-11 |
| d3,VP | d6,VP | –0.542 | –0.344 | –0.145 | 4.09E-05 |
| d1,D2 OT | d1,D1 OT | –0.170 | 0.011 | 0.192 | 1.000 |
| d1,D2 OT | d1,VP | –0.141 | 0.049 | 0.239 | 0.995 |
| d1,D1 OT | d1,VP | –0.152 | 0.038 | 0.228 | 0.999 |
| d3,D2 OT | d3,D1 OT | –0.204 | –0.023 | 0.158 | 1.000 |
| d3,D2 OT | d3,VP | –0.341 | –0.151 | 0.039 | 0.221 |
| d3,D1 OT | d3,VP | –0.318 | –0.128 | 0.062 | 0.427 |
| d6,D2 OT | d6,D1 OT | –0.309 | –0.127 | 0.054 | 0.370 |
| d6,D2 OT | d6,VP | –0.665 | –0.475 | –0.285 | 1.15E-08 |
| d6,D1 OT | d6,VP | –0.538 | –0.348 | –0.158 | 1.43E-05 |
Two-way ANOVA for effect of day or lens placement on percentage of neurons with auROC >0.75 for {SK vs. XK} (Figure 3J).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| day | 0.551 | 2 | 0.275 | 28.099 | 1.794E-08 |
| region | 0.377 | 2 | 0.188 | 19.265 | 1.156E-06 |
| day:region | 0.364 | 4 | 0.0912 | 9.310 | 1.764E-05 |
| Error | 0.411 | 42 | 0.009 | ||
| Total | 1.637 | 50 |
Post hoc comparisons of percentage of neurons with auROC >0.75 for {SK vs. XK} across imaging day and region (Figure 3J).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,D2 OT | d3,D2 OT | –0.189 | –0.002 | 0.184 | 1.000 |
| d1,D2 OT | d6,D2 OT | –0.238 | –0.052 | 0.135 | 0.992 |
| d3,D2 OT | d6,D2 OT | –0.236 | –0.049 | 0.137 | 0.994 |
| d1,D1 OT | d3,D1 OT | –0.313 | –0.126 | 0.061 | 0.421 |
| d1,D1 OT | d6,D1 OT | –0.356 | –0.170 | 0.017 | 0.102 |
| d3,D1 OT | d6,D1 OT | –0.230 | –0.044 | 0.143 | 0.997 |
| d1,VP | d3,VP | –0.454 | –0.250 | –0.045 | 7.23E-03 |
| d1,VP | d6,VP | –0.750 | –0.545 | –0.341 | 2.05E-09 |
| d3,VP | d6,VP | –0.500 | –0.295 | –0.091 | 8.14E-04 |
| d1,D2 OT | d1,D1 OT | –0.219 | –0.033 | 0.154 | 1.000 |
| d1,D2 OT | d1,VP | –0.163 | 0.033 | 0.229 | 1.000 |
| d1,D1 OT | d1,VP | –0.130 | 0.066 | 0.261 | 0.972 |
| d3,D2 OT | d3,D1 OT | –0.343 | –0.156 | 0.031 | 0.167 |
| d3,D2 OT | d3,VP | –0.410 | –0.214 | –0.019 | 2.26E-02 |
| d3,D1 OT | d3,VP | –0.254 | –0.058 | 0.138 | 0.987 |
| d6,D2 OT | d6,D1 OT | –0.337 | –0.151 | 0.036 | 0.204 |
| d6,D2 OT | d6,VP | –0.656 | –0.461 | –0.265 | 5.39E-08 |
| d6,D1 OT | d6,VP | –0.506 | –0.310 | –0.114 | 1.95E-04 |
Two-way ANOVA for effect of day or lens placement on percentage of neurons with auROC >0.75 for {SK vs. ST} (Figure 3K).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| day | 0.01400 | 2 | 0.00700 | 0.45251 | 0.63909 |
| region | 0.08031 | 2 | 0.04015 | 2.59595 | 0.08650 |
| day:region | 0.01282 | 4 | 0.00321 | 0.20726 | 0.93297 |
| Error | 0.64967 | 42 | 0.01547 | ||
| Total | 0.75527 | 50 |
Post hoc comparisons of percentage of neurons with auROC >0.75 for {SK vs. ST} across imaging day and region (Figure 3K).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,D2 OT | d3,D2 OT | –0.237 | –0.002 | 0.232 | 1.000 |
| d1,D2 OT | d6,D2 OT | –0.276 | –0.041 | 0.193 | 1.000 |
| d3,D2 OT | d6,D2 OT | –0.274 | –0.039 | 0.196 | 1.000 |
| d1,D1 OT | d3,D1 OT | –0.255 | –0.020 | 0.214 | 1.000 |
| d1,D1 OT | d6,D1 OT | –0.233 | 0.001 | 0.236 | 1.000 |
| d3,D1 OT | d6,D1 OT | –0.213 | 0.022 | 0.257 | 1.000 |
| d1,VP | d3,VP | –0.316 | –0.059 | 0.198 | 0.998 |
| d1,VP | d6,VP | –0.336 | –0.079 | 0.178 | 0.983 |
| d3,VP | d6,VP | –0.277 | –0.020 | 0.237 | 1.000 |
| d1,D2 OT | d1,D1 OT | –0.267 | –0.032 | 0.202 | 1.000 |
| d1,D2 OT | d1,VP | –0.142 | 0.104 | 0.350 | 0.900 |
| d1,D1 OT | d1,VP | –0.110 | 0.136 | 0.382 | 0.678 |
| d3,D2 OT | d3,D1 OT | –0.285 | –0.050 | 0.184 | 0.999 |
| d3,D2 OT | d3,VP | –0.199 | 0.047 | 0.293 | 0.999 |
| d3,D1 OT | d3,VP | –0.149 | 0.097 | 0.344 | 0.928 |
| d6,D2 OT | d6,D1 OT | –0.224 | 0.011 | 0.245 | 1.000 |
| d6,D2 OT | d6,VP | –0.180 | 0.066 | 0.312 | 0.993 |
| d6,D1 OT | d6,VP | –0.191 | 0.055 | 0.302 | 0.998 |
Pairwise comparisons of |ΔΔFday3|-|ΔΔFday1| across regions (Figure 4I).
| Group A | Group B | Lower Limit | A-B | Upper Limit | FDR adjusted p-value |
|---|---|---|---|---|---|
| D2 OT | D1 OT | –0.204 | –0.138 | –0.073 | 4.37E-02 |
| D2 OT | VP | –0.275 | –0.214 | –0.153 | 1.08E-03 |
| D1 OT | VP | –0.120 | –0.075 | –0.031 | 0.141 |
One sample t-tests of |ΔΔFday3|-|ΔΔFday1| in different regions (Figure 4I).
| Population | Lower Limit | Mean | Upper Limit | FDR adjusted p-value |
|---|---|---|---|---|
| D2 OT | –0.146 | –6.51E-02 | 1.58E-02 | 0.259 |
| D1 OT | 2.78E-02 | 7.83E-02 | 0.129 | 4.48E-02 |
| VP | 0.133 | 0.174 | 0.215 | 2.49E-07 |
One-way ANOVA for effect of region on {S vs. X|P} linear classifier accuracy (Figure 5G).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.018 | 2 | 9.10E-03 | 9.569 | 2.40E-03 |
| Error | 0.013 | 14 | 9.51E-04 | ||
| Total | 0.032 | 16 |
Post hoc comparisons of {S vs. X|P} linear classifier accuracy across regions (Figure 5G).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT | D1 OT | –0.114 | –0.067 | –0.020 | 5.57E-03 |
| D2 OT | VP | –0.119 | –0.070 | –0.021 | 5.65E-03 |
| D1 OT | VP | –0.052 | –0.003 | 0.046 | 0.985 |
One-way ANOVA for effect of region on generalized {S vs. X|P} linear classifier accuracy (Figure 5G).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.134 | 2 | 0.067 | 14.136 | 4.37E-04 |
| Error | 0.066 | 14 | 0.005 | ||
| Total | 0.201 | 16 |
Post hoc comparisons of generalized {S vs. X|P} linear classifier accuracy across regions (Figure 5G).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT | D1 OT | –0.153 | –0.049 | 0.055 | 0.451 |
| D2 OT | VP | –0.323 | –0.214 | –0.105 | 4.17E-04 |
| D1 OT | VP | –0.274 | –0.165 | –0.056 | 3.84E-03 |
Two-way ANOVA for effect of imaging days or region on normalized PR (Figure 5I).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| days | 0.775 | 2 | 0.387 | 0.277 | 0.759 |
| region | 50.226 | 2 | 25.113 | 17.969 | 2.704E-06 |
| days:region | 14.484 | 4 | 3.621 | 2.591 | 0.051 |
Post hoc comparisons of normalized PR across imaging day and region (Figure 5I).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,D2 OT | d3,D2 OT | –2.938 | –0.592 | 1.754 | 0.995 |
| d1,D2 OT | d6,D2 OT | –3.700 | –1.355 | 0.991 | 0.623 |
| d3,D2 OT | d6,D2 OT | –2.999 | –0.762 | 1.474 | 0.968 |
| d1,D1 OT | d3,D1 OT | –1.948 | 0.289 | 2.526 | 1 |
| d1,D1 OT | d6,D1 OT | –1.965 | 0.272 | 2.509 | 1 |
| d3,D1 OT | d6,D1 OT | –2.254 | –0.017 | 2.220 | 1 |
| d1,VP | d3,VP | –2.404 | 0.195 | 2.794 | 1 |
| d1,VP | d6,VP | –0.783 | 1.816 | 4.415 | 0.372 |
| d3,VP | d6,VP | –0.829 | 1.621 | 4.071 | 0.445 |
| d1,D2 OT | d3,D1 OT | –3.316 | –0.971 | 1.375 | 0.907 |
| d1,D2 OT | d1,VP | –1.921 | 0.678 | 3.277 | 0.994 |
| d1,D1 OT | d1,VP | –0.563 | 1.938 | 4.438 | 0.245 |
| d3,D2 OT | d3,D1 OT | –2.615 | –0.378 | 1.858 | 1 |
| d3,D2 OT | d3,VP | –0.881 | 1.465 | 3.811 | 0.522 |
| d3,D1 OT | d3,VP | –0.502 | 1.843 | 4.189 | 0.229 |
| d6,D2 OT | d6,D1 OT | –1.870 | 0.367 | 2.604 | 1 |
| d6,D2 OT | d6,VP | 1.503 | 3.848 | 6.194 | 1.14E-04 |
| d6,D1 OT | d6,VP | 1.135 | 3.481 | 5.827 | 5.67E-04 |
One-way ANOVA for effect of region on {SK vs. PK} linear classifier accuracy trained on PC1 (Figure 5L).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.209 | 2 | 0.104 | 20.965 | 6.16E-05 |
| Error | 0.070 | 14 | 0.005 | ||
| Total | 0.279 | 16 |
Post hoc comparisons of {SK vs. PK} linear classifier accuracy trained on PC1 across regions (Figure 5L).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT | D1 OT | –0.172 | –0.066 | 0.041 | 0.273 |
| D2 OT | VP | –0.380 | –0.268 | –0.157 | 5.64E-05 |
| D1 OT | VP | –0.315 | –0.203 | –0.091 | 8.57E-04 |
One-way ANOVA for effect of region on {SK vs. ST} linear classifier accuracy trained on PC1-PC15 (Figure 5L).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.019 | 2 | 0.009 | 0.646 | 0.539 |
| Error | 0.206 | 14 | 0.015 | ||
| Total | 0.225 | 16 |
Post hoc comparisons of {SK vs. ST} linear classifier accuracy trained on PC1-PC15 across regions (Figure 5L).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT | D1 OT | –0.203 | –0.019 | 0.164 | 0.958 |
| D2 OT | VP | –0.131 | 0.061 | 0.253 | 0.687 |
| D1 OT | VP | –0.111 | 0.081 | 0.273 | 0.529 |
Two-way ANOVA for effect of lick spout presence and sucrose contingency on anticipatory licking (Figure 6C).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| spout | 17.176 | 1 | 17.176 | 50.125 | 2.56E-07 |
| S% | 19.415 | 2 | 9.707 | 28.329 | 4.82E-07 |
| spout:S% | 11.533 | 2 | 5.767 | 16.829 | 2.71E-05 |
| Error | 8.224 | 24 | 0.343 | ||
| Total | 56.348 | 29 |
Post hoc comparisons of anticipatory licking across spout presence and sucrose contingency (Figure 6C).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| spout = 0, S0.0 | spout = 1, S0.0 | –1.205 | –0.060 | 1.085 | 1 |
| spout = 0, S0.0 | spout = 0, S0.5 | –1.335 | –0.190 | 0.955 | 0.995 |
| spout = 0, S0.0 | spout = 1, S0.5 | –2.725 | –1.580 | –0.435 | 3.24E-03 |
| spout = 0, S0.0 | spout = 0, S1.0 | –1.595 | –0.450 | 0.695 | 0.825 |
| spout = 0, S0.0 | spout = 1, S1.0 | –4.685 | –3.540 | –2.395 | 1.66E-08 |
| spout = 1, S0.0 | spout = 0, S0.5 | –1.275 | –0.130 | 1.015 | 0.999 |
| spout = 1, S0.0 | spout = 1, S0.5 | –2.665 | –1.520 | –0.375 | 4.80E-03 |
| spout = 1, S0.0 | spout = 0, S1.0 | –1.535 | –0.390 | 0.755 | 0.895 |
| spout = 1, S0.0 | spout = 1, S1.0 | –4.625 | –3.480 | –2.335 | 2.29E-08 |
| spout = 0, S0.5 | spout = 1, S0.5 | –2.535 | –1.390 | –0.245 | 1.11E-02 |
| spout = 0, S0.5 | spout = 0, S1.0 | –1.405 | –0.260 | 0.885 | 0.980 |
| spout = 0, S0.5 | spout = 1, S1.0 | –4.495 | –3.350 | –2.205 | 4.70E-08 |
| spout = 1, S0.5 | spout = 0, S1.0 | –0.015 | 1.130 | 2.275 | 5.44E-02 |
| spout = 1, S0.5 | spout = 1, S1.0 | –3.105 | –1.960 | –0.815 | 2.58E-04 |
| spout = 0, S1.0 | spout = 1, S1.0 | –4.235 | –3.090 | –1.945 | 2.07E-07 |
Two-way ANOVA for effect of imaging day and valence of odor on velocity during cue presentation (Figure 2—figure supplement 5E).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| days | 466.659 | 5 | 93.332 | 1.199 | 0.308 |
| valence | 217.697 | 2 | 108.849 | 1.399 | 0.248 |
| days:valence | 671.044 | 10 | 67.104 | 0.862 | 0.569 |
| Error | 35948.767 | 462 | 77.811 | ||
| Total | 37302.764 | 479 |
Two-way ANOVA for effect of imaging day and valence of odor on velocity during unconditioned stimulus (Figure 2—figure supplement 5E).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| days | 851.615 | 5 | 170.323 | 0.660 | 0.654 |
| valence | 29846.379 | 2 | 14923.189 | 57.811 | 3.91E-23 |
| days:valence | 2979.836 | 10 | 297.984 | 1.154 | 0.320 |
| Error | 119259.047 | 462 | 258.136 | ||
| Total | 153052.200 | 479 |
Post hoc comparisons of velocity during unconditioned stimulus across imaging days and valence of odor (Figure 2—figure supplement 5E).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| d1,P | d1,X | 5.990 | 20.970 | 35.951 | 1.51E-04 |
| d1,P | d1,S | 10.286 | 25.266 | 40.246 | 5.70E-07 |
| d2,P | d2,X | –3.390 | 11.591 | 26.571 | 0.381 |
| d2,P | d2,S | –2.310 | 12.670 | 27.651 | 0.225 |
| d3,P | d3,X | –4.604 | 10.942 | 26.488 | 0.565 |
| d3,P | d3,S | –1.079 | 14.466 | 30.012 | 0.104 |
| d4,P | d4,X | –4.253 | 11.292 | 26.838 | 0.504 |
| d4,P | d4,S | –1.729 | 13.817 | 29.363 | 0.155 |
| d5,P | d5,X | –2.610 | 12.936 | 28.482 | 0.251 |
| d5,P | d5,S | 4.085 | 19.631 | 35.177 | 1.45E-03 |
| d6,P | d6,X | –1.619 | 13.927 | 29.473 | 0.145 |
| d6,P | d6,S | 10.737 | 26.283 | 41.829 | 5.22E-07 |
| d1,P | d2,P | –5.400 | 9.580 | 24.560 | 0.733 |
| d1,P | d3,P | –5.063 | 10.203 | 25.469 | 0.660 |
| d1,P | d4,P | –4.305 | 10.961 | 26.227 | 0.526 |
| d1,P | d5,P | –6.954 | 8.311 | 23.577 | 0.913 |
| d1,P | d6,P | –10.637 | 4.628 | 19.894 | 1 |
| d2,P | d3,P | –14.643 | 0.623 | 15.889 | 1 |
| d2,P | d4,P | –13.885 | 1.381 | 16.647 | 1 |
| d2,P | d5,P | –16.534 | –1.269 | 13.997 | 1 |
| d2,P | d6,P | –20.217 | –4.951 | 10.314 | 1 |
| d3,P | d4,P | –14.788 | 0.758 | 16.304 | 1 |
| d3,P | d5,P | –17.437 | –1.892 | 13.654 | 1 |
| d3,P | d6,P | –21.120 | –5.574 | 9.971 | 0.999 |
| d4,P | d5,P | –18.196 | –2.650 | 12.896 | 1 |
| d4,P | d6,P | –21.878 | –6.333 | 9.213 | 0.995 |
| d5,P | d6,P | –19.229 | –3.683 | 11.863 | 1 |
| d1,X | d2,X | –14.780 | 0.200 | 15.181 | 1 |
| d1,X | d3,X | –15.091 | 0.175 | 15.441 | 1 |
| d1,X | d4,X | –13.982 | 1.283 | 16.549 | 1 |
| d1,X | d5,X | –14.989 | 0.277 | 15.543 | 1 |
| d1,X | d6,X | –17.680 | –2.414 | 12.851 | 1 |
| d2,X | d3,X | –15.291 | –0.025 | 15.240 | 1 |
| d2,X | d4,X | –14.183 | 1.083 | 16.349 | 1 |
| d2,X | d5,X | –15.189 | 0.077 | 15.342 | 1 |
| d2,X | d6,X | –17.881 | –2.615 | 12.651 | 1 |
| d3,X | d4,X | –14.437 | 1.108 | 16.654 | 1 |
| d3,X | d5,X | –15.444 | 0.102 | 15.648 | 1 |
| d3,X | d6,X | –18.135 | –2.589 | 12.956 | 1 |
| d4,X | d5,X | –16.552 | –1.006 | 14.540 | 1 |
| d4,X | d6,X | –19.244 | –3.698 | 11.848 | 1 |
| d5,X | d6,X | –18.237 | –2.691 | 12.854 | 1 |
| d1,S | d2,S | –17.996 | –3.015 | 11.965 | 1 |
| d1,S | d3,S | –15.862 | –0.597 | 14.669 | 1 |
| d1,S | d4,S | –15.753 | –0.488 | 14.778 | 1 |
| d1,S | d5,S | –12.589 | 2.677 | 17.942 | 1 |
| d1,S | d6,S | –9.620 | 5.646 | 20.911 | 0.998 |
| d2,S | d3,S | –12.847 | 2.419 | 17.685 | 1 |
| d2,S | d4,S | –12.738 | 2.528 | 17.794 | 1 |
| d2,S | d5,S | –9.574 | 5.692 | 20.958 | 0.998 |
| d2,S | d6,S | –6.605 | 8.661 | 23.927 | 0.880 |
| d3,S | d4,S | –15.437 | 0.109 | 15.655 | 1 |
| d3,S | d5,S | –12.273 | 3.273 | 18.819 | 1 |
| d3,S | d6,S | –9.304 | 6.242 | 21.788 | 0.996 |
| d4,S | d5,S | –12.382 | 3.164 | 18.710 | 1 |
| d4,S | d6,S | –9.413 | 6.133 | 21.679 | 0.997 |
| d5,S | d6,S | –12.577 | 2.969 | 18.515 | 1 |
Two-way ANOVA for effect of imaging day and valence of odor on relative eye size during cue presentation (Figure 2—figure supplement 5G).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| days | 0.293 | 5 | 0.059 | 12.301 | 8.84E-11 |
| valence | 0.014 | 2 | 0.007 | 1.523 | 0.220 |
| days:valence | 0.121 | 10 | 0.012 | 2.546 | 5.95E-03 |
| Error | 1.313 | 276 | 0.005 | ||
| Total | 1.739 | 293 |
Two-way ANOVA for effect of imaging day and valence of odor on relative eye size during unconditioned stimulus (Figure 2—figure supplement 5G).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| days | 0.178 | 5 | 0.036 | 4.529 | 5.51E-04 |
| valence | 0.040 | 2 | 0.020 | 2.574 | 0.078 |
| days:valence | 0.167 | 10 | 0.017 | 2.123 | 2.29E-02 |
| Error | 2.167 | 276 | 0.008 | ||
| Total | 2.547 | 293 |
Four-way ANOVA for effect of imaging day, valence, functional group, and region on the percentage of neurons responsive to a given odor (Figure 2—figure supplement 7A).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| ket | 0.051 | 1 | 0.051 | 3.828 | 0.051 |
| val. | 0.787 | 2 | 0.393 | 29.766 | 2.48E-12 |
| reg. | 6.62E-03 | 2 | 0.003 | 0.250 | 0.779 |
| day | 0.928 | 2 | 0.464 | 35.101 | 3.57E-14 |
| ket:val. | 0.024 | 2 | 0.012 | 0.911 | 0.403 |
| ket:reg. | 0.018 | 2 | 0.009 | 0.671 | 0.512 |
| ket:day | 0.057 | 2 | 0.029 | 2.163 | 0.117 |
| val.:reg. | 0.622 | 4 | 0.155 | 11.763 | 8.94E-09 |
| val.:day | 0.264 | 4 | 0.066 | 4.995 | 6.85E-04 |
| reg.:day | 0.328 | 4 | 0.082 | 6.212 | 8.79E-05 |
| ket:val.:reg. | 0.054 | 4 | 0.014 | 1.025 | 0.395 |
| ket:val.:day | 0.095 | 4 | 0.024 | 1.796 | 0.130 |
| ket:reg.:day | 0.014 | 4 | 0.004 | 0.272 | 0.896 |
| val.:reg.:day | 0.241 | 8 | 0.030 | 2.277 | 0.023 |
| ket:val.:reg.: day | 0.062 | 8 | 0.008 | 0.582 | 0.792 |
| Error | 3.331 | 252 | 0.013 | ||
| Total | 6.670 | 305 |
Linear model of the fixed effects of region, imaging day, and valence and the random effect of individual animal on |ΔΔF/F| (Figure 2—figure supplement 8A).
| Formula: | |||||
|---|---|---|---|---|---|
| Fmag ~1 + reg*day +reg*val +day*val +reg:day:val + (1 | id) | |||||
| Model information | |||||
| # of observations: | Fixed effects coefficients: | Random effect coefficients: | Covariance parameters: | ||
| 11,160 | 12 | 17 | 2 | ||
| Model fit statistics: | |||||
| AIC | BIC | Log Likelihood | Deviance | ||
| 9050.1 | 9152.6 | –4511.1 | 9022.1 | ||
| Fixed effects coefficients (95% CIs): | |||||
| Name | Estimate | SE | tStat | DF | pValue |
| intercept | 0.469 | 0.0155 | 30.228 | 11,148 | 5.43E-193 |
| reg_D1 | –0.0222 | 0.0205 | –1.0829 | 11,148 | 0.279 |
| reg_VP | 0.0192 | 0.0255 | 0.753 | 11,148 | 0.452 |
| day | –0.0140 | 0.00707 | –1.973 | 11,148 | 0.0485 |
| val | –0.0244 | 0.0190 | –1.286 | 11,148 | 0.198 |
| reg_D1:day | 0.00806 | 0.00947 | 0.852 | 11,148 | 0.394 |
| reg_VP:day | –0.0101 | 0.0117 | –0.862 | 11,148 | 0.389 |
| reg_D1:val | –0.00659 | 0.0251 | –0.262 | 11,148 | 0.793 |
| reg_VP:val | –0.0359 | 0.0312 | –1.150 | 11,148 | 0.250 |
| day:val | 0.00799 | 0.00866 | 0.922 | 11,148 | 0.356 |
| reg_D1:day:val | 0.0294 | 0.0116 | 2.539 | 11,148 | 0.0111 |
| reg_VP:day:val | 0.0826 | 0.0143 | 5.762 | 11,148 | 8.5E-9 |
Linear model of the fixed effects of region and imaging day, and the random effect of individual animals on the auROC of single-neuron {S vs. X|P} classifiers (Figure 3—figure supplement 1F).
| Formula: | |||||
|---|---|---|---|---|---|
| auROC {S vs. X|P}~1 + region*day + (1 | id) | |||||
| Model information | |||||
| Number of observations: | Fixed effects coefficients: | Random effect coefficients: | Covariance parameters: | ||
| 1860 | 6 | 17 | 2 | ||
| Model fit statistics: | |||||
| AIC | BIC | Log Likelihood | Deviance | ||
| –4303.7 | –4259.5 | 2159.8 | –4319.7 | ||
| Fixed effects coefficients (95% CIs): | |||||
| Name | Estimate | SE | tStat | DF | pValue |
| intercept | 0.617 | 0.009 | 68.007 | 1854 | 0 |
| reg_D1 | –0.002 | 0.012 | –0.186 | 1854 | 0.853 |
| reg_VP | –0.037 | 0.014 | –2.665 | 1854 | 7.76E-03 |
| day | 0.003 | 0.001 | 1.874 | 1854 | 0.061 |
| reg_D1:day | 0.007 | 0.002 | 3.484 | 1854 | 5.06E-04 |
| reg_VP:day | 0.029 | 0.002 | 12.056 | 1854 | 2.80E-32 |
Two-way ANOVA for effect of imaging day and region on the median auROC value of {S vs. X|P} classifiers for each animal (Figure 3—figure supplement 1F).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.030 | 2 | 0.015 | 14.686 | 1.46E-05 |
| day | 0.050 | 2 | 0.025 | 24.336 | 9.58E-08 |
| region:day | 0.041 | 4 | 0.010 | 10.001 | 8.88E-06 |
| Error | 0.043 | 42 | 0.001 | ||
| Total | 0.158 | 50 |
Post hoc comparison of the median auROC value for {S vs. X|P} across imaging day and region (Figure 3—figure supplement 1F).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT,d1 | D2 OT,d3 | –0.072 | –0.012 | 0.049 | 0.999 |
| D2 OT,d1 | D2 OT,d6 | –0.075 | –0.014 | 0.046 | 0.997 |
| D2 OT,d3 | D2 OT,d6 | –0.063 | –0.003 | 0.058 | 1 |
| D1 OT,d3 | D1 OT,d6 | –0.072 | –0.012 | 0.049 | 0.999 |
| D1 OT,d1 | D1 OT,d3 | –0.091 | –0.030 | 0.030 | 0.779 |
| D1 OT,d1 | D1 OT,d6 | –0.102 | –0.042 | 0.019 | 0.387 |
| VP,d1 | VP,d3 | –0.130 | –0.064 | 0.002 | 6.62E-02 |
| VP,d1 | VP,d6 | –0.241 | –0.174 | –0.108 | 2.83E-09 |
| VP,d3 | VP,d6 | –0.177 | –0.111 | –0.044 | 7.91E-05 |
| D2 OT,d1 | D1 OT,d1 | –0.056 | 0.005 | 0.065 | 1 |
| D2 OT,d1 | VP,d1 | –0.050 | 0.013 | 0.076 | 0.999 |
| D1 OT,d1 | VP,d1 | –0.055 | 0.008 | 0.072 | 1 |
| D2 OT,d3 | D1 OT,d3 | –0.074 | –0.014 | 0.047 | 0.998 |
| D2 OT,d3 | VP,d3 | –0.103 | –0.039 | 0.024 | 0.537 |
| D1 OT,d3 | VP,d3 | –0.089 | –0.026 | 0.038 | 0.921 |
| D2 OT,d6 | D1 OT,d6 | –0.083 | –0.023 | 0.038 | 0.948 |
| D2 OT,d6 | VP,d6 | –0.210 | –0.147 | –0.084 | 7.68E-08 |
| D1 OT,d6 | VP,d6 | –0.188 | –0.124 | –0.061 | 3.44E-06 |
Linear model of the fixed effects of region and imaging day, and the random effect of individual animals on the auROC of single-neuron {SK vs. ST} classifiers (Figure 3—figure supplement 1G).
| Formula: | |||||
|---|---|---|---|---|---|
| auROC {SK vs. ST}~1 + region*day + (1 | id) | |||||
| Model information | |||||
| Number of observations: | Fixed effects coefficients: | Random effect coefficients: | Covariance parameters: | ||
| 1860 | 6 | 17 | 2 | ||
| Model fit statistics: | |||||
| AIC | BIC | Log Likelihood | Deviance | ||
| –2908.8 | –2864.5 | 1462.4 | –2924.8 | ||
| Fixed effects coefficients (95% CIs): | |||||
| Name | Estimate | SE | tStat | DF | pValue |
| intercept | 0.636 | 0.015 | 42.040 | 1854 | 7.72E-272 |
| region_D1 | –0.020 | 0.021 | –0.962 | 1854 | 0.336 |
| region_VP | –0.024 | 0.024 | –1.014 | 1854 | 0.311 |
| day | 9.67E-04 | 5.25E-03 | 0.184 | 1854 | 0.854 |
| region_D1:day | 0.011 | 0.007 | 1.622 | 1854 | 0.105 |
| region_VP:day | –0.003 | 0.009 | –0.346 | 1854 | 0.730 |
Two-way ANOVA for effect of imaging day and region on the median auROC value of {SK vs. ST} classifiers for each animal (Figure 3—figure supplement 1G).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.011 | 2 | 5.54E-03 | 2.793 | 0.073 |
| day | 2.91E-04 | 2 | 1.46E-04 | 0.073 | 0.929 |
| region:day | 3.66E-03 | 4 | 9.16E-04 | 0.462 | 0.763 |
| Error | 0.083 | 42 | 1.98E-03 | ||
| Total | 0.098 | 50 |
Linear model of the fixed effects of region and imaging day, and the random effect of individual animals on the single-neuron valence scores (Figure 3—figure supplement 1H).
| Formula: | |||||
|---|---|---|---|---|---|
| score ~1 + region*day + (1 | id) | |||||
| Model information | |||||
| Number of observations: | Fixed effects coefficients: | Random effect coefficients: | Covariance parameters: | ||
| 1860 | 6 | 17 | 2 | ||
| Model fit statistics: | |||||
| AIC | BIC | Log Likelihood | Deviance | ||
| –3219.9 | –3175.7 | 1618 | –3235.9 | ||
| Fixed effects coefficients (95% CIs): | |||||
| Name | Estimate | SE | tStat | DF | pValue |
| intercept | –0.023 | 0.015 | –1.563 | 1854 | 0.118 |
| region_D1 | 4.19E-03 | 0.020 | 0.204 | 1854 | 0.838 |
| region_VP | –0.062 | 0.023 | –2.648 | 1854 | 8.16E-03 |
| day | 5.84E-03 | 4.83E-03 | 1.209 | 1854 | 0.227 |
| region_D1:day | 6.40E-03 | 6.45E-03 | 0.991 | 1854 | 0.322 |
| region_VP:day | 0.075 | 7.98E-03 | 9.384 | 1854 | 1.80E-20 |
Two-way ANOVA for effect of imaging day and region on the median valence score for each animal (Figure 3—figure supplement 1H).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| reg | 0.070 | 2 | 0.035 | 13.163 | 3.65E-05 |
| day | 0.032 | 2 | 0.016 | 6.073 | 4.82E-03 |
| reg:day | 0.045 | 4 | 0.011 | 4.264 | 5.50E-03 |
| Error | 0.111 | 42 | 2.65E-03 | ||
| Total | 0.253 | 50 |
Post hoc comparison of the median valence scores across imaging day and region (Figure 3—figure supplement 1H).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT,d1 | D2 OT,d3 | –0.100 | –0.003 | 0.094 | 1 |
| D2 OT,d1 | D2 OT,d6 | –0.109 | –0.012 | 0.086 | 1 |
| D2 OT,d3 | D2 OT,d6 | –0.106 | –0.008 | 0.089 | 1 |
| D1 OT,d1 | D1 OT,d3 | –0.100 | –0.002 | 0.095 | 1 |
| D1 OT,d1 | D1 OT,d6 | –0.102 | –0.005 | 0.092 | 1 |
| D1 OT,d3 | D1 OT,d6 | –0.100 | –0.003 | 0.095 | 1 |
| VP,d1 | VP,d3 | –0.157 | –0.051 | 0.056 | 0.819 |
| VP,d1 | VP,d6 | –0.271 | –0.165 | –0.058 | 2.87E-04 |
| VP,d3 | VP,d6 | –0.220 | –0.114 | –0.007 | 2.87E-02 |
| D2 OT,d1 | D1 OT,d1 | –0.112 | –0.015 | 0.082 | 1 |
| D2 OT,d1 | VP,d1 | –0.122 | –0.020 | 0.082 | 1 |
| D1 OT,d1 | VP,d1 | –0.107 | –0.005 | 0.097 | 1 |
| D2 OT,d3 | D1 OT,d3 | –0.111 | –0.014 | 0.083 | 1 |
| D2 OT,d3 | VP,d3 | –0.169 | –0.067 | 0.034 | 0.447 |
| D1 OT,d3 | VP,d3 | –0.155 | –0.053 | 0.049 | 0.736 |
| D2 OT,d6 | D1 OT,d6 | –0.105 | –0.008 | 0.089 | 1 |
| D2 OT,d6 | VP,d6 | –0.275 | –0.173 | –0.071 | 6.07E-05 |
| D1 OT,d6 | VP,d6 | –0.266 | –0.164 | –0.062 | 1.41E-04 |
Two-way ANOVA for effect of imaging day and region on the median single-neuron MNR accuracy for each animal Figure 3—figure supplement 2B.
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| reg | 0.003 | 2 | 0.002 | 4.286 | 2.02E-02 |
| day | 0.006 | 2 | 0.003 | 7.181 | 2.08E-03 |
| reg:day | 0.004 | 4 | 0.001 | 2.622 | 4.82E-02 |
| Error | 0.017 | 42 | 0.000 | ||
| Total | 0.030 | 50 |
Post hoc comparison of median single-neuron MNR accuracy across imaging day and region (Figure 3—figure supplement 2B).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT,d1 | D2 OT,d3 | –0.051 | –0.013 | 0.024 | 0.960 |
| D2 OT,d1 | D2 OT,d6 | –0.045 | –0.007 | 0.031 | 0.999 |
| D2 OT,d3 | D2 OT,d6 | –0.032 | 0.006 | 0.044 | 1 |
| D1 OT,d1 | D1 OT,d3 | –0.047 | –0.010 | 0.028 | 0.996 |
| D1 OT,d1 | D1 OT,d6 | –0.050 | –0.012 | 0.026 | 0.981 |
| D1 OT,d3 | D1 OT,d6 | –0.040 | –0.002 | 0.036 | 1 |
| VP,d1 | VP,d3 | –0.073 | –0.031 | 0.011 | 0.294 |
| VP,d1 | VP,d6 | –0.099 | –0.058 | –0.016 | 1.47E-03 |
| VP,d3 | VP,d6 | –0.068 | –0.027 | 0.015 | 0.490 |
| D2 OT,d1 | D1 OT,d1 | –0.052 | –0.014 | 0.024 | 0.953 |
| D2 OT,d1 | VP,d1 | –0.037 | 0.003 | 0.043 | 1 |
| D1 OT,d1 | VP,d1 | –0.023 | 0.017 | 0.057 | 0.896 |
| D2 OT,d3 | D1 OT,d3 | –0.048 | –0.010 | 0.028 | 0.994 |
| D2 OT,d3 | VP,d3 | –0.054 | –0.014 | 0.025 | 0.955 |
| D1 OT,d3 | VP,d3 | –0.044 | –0.005 | 0.035 | 1 |
| D2 OT,d6 | D1 OT,d6 | –0.057 | –0.019 | 0.019 | 0.797 |
| D2 OT,d6 | VP,d6 | –0.087 | –0.047 | –0.008 | 9.47E-03 |
| D1 OT,d6 | VP,d6 | –0.069 | –0.029 | 0.011 | 0.329 |
Two-way ANOVA for effect of imaging day and region on the median single-neuron MNR shuffled accuracy for each animal Figure 3—figure supplement 2B.
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| reg | 2.04E-08 | 2 | 1.02E-08 | 0.040 | 0.961 |
| day | 7.71E-07 | 2 | 3.86E-07 | 1.519 | 0.231 |
| reg:day | 1.22E-06 | 4 | 3.05E-07 | 1.200 | 0.325 |
| Error | 1.07E-05 | 42 | 2.54E-07 | ||
| Total | 1.26E-05 | 50 |
Two-way ANOVA for effect of imaging day and region on the median S-cue/S-cue confusion for each animal Figure 3—figure supplement 2D.
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| reg | 0.074 | 2 | 0.037 | 27.720 | 2.11E-08 |
| day | 0.019 | 2 | 9.43E-03 | 7.066 | 2.26E-03 |
| reg:day | 0.011 | 4 | 2.65E-03 | 1.986 | 1.14E-01 |
| Error | 0.056 | 42 | 1.33E-03 | ||
| Total | 0.157 | 50 |
Post hoc comparison of median S-cue/S-cue confusion across imaging day and region Figure 3—figure supplement 2D.
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT,d1 | D1 OT,d1 | –0.090 | –0.021 | 0.048 | 0.985 |
| D2 OT,d1 | VP,d1 | –0.132 | –0.060 | 0.012 | 0.174 |
| D1 OT,d1 | VP,d1 | –0.111 | –0.039 | 0.033 | 0.700 |
| D2 OT,d3 | D1 OT,d3 | –0.095 | –0.026 | 0.043 | 0.940 |
| D2 OT,d3 | VP,d3 | –0.156 | –0.084 | –0.011 | 1.30E-02 |
| D1 OT,d3 | VP,d3 | –0.130 | –0.057 | 0.015 | 0.223 |
| D2 OT,d6 | D1 OT,d6 | –0.084 | –0.015 | 0.054 | 0.998 |
| D2 OT,d6 | VP,d6 | –0.204 | –0.132 | –0.059 | 1.55E-05 |
| D1 OT,d6 | VP,d6 | –0.189 | –0.116 | –0.044 | 1.46E-04 |
| D2 OT,d1 | D2 OT,d3 | –0.079 | –0.010 | 0.059 | 1 |
| D2 OT,d1 | D2 OT,d6 | –0.094 | –0.025 | 0.044 | 0.955 |
| D2 OT,d3 | D2 OT,d6 | –0.084 | –0.015 | 0.054 | 0.998 |
| D1 OT,d1 | D1 OT,d3 | –0.084 | –0.015 | 0.054 | 0.998 |
| D1 OT,d1 | D1 OT,d6 | –0.088 | –0.019 | 0.049 | 0.990 |
| D1 OT,d3 | D1 OT,d6 | –0.073 | –0.004 | 0.065 | 1 |
| VP,d1 | VP,d3 | –0.109 | –0.033 | 0.042 | 0.874 |
| VP,d1 | VP,d6 | –0.172 | –0.097 | –0.021 | 4.11E-03 |
| VP,d3 | VP,d6 | –0.139 | –0.063 | 0.012 | 0.165 |
Two-way ANOVA for effect of imaging day and region on the median confusion within functional groups for each animal Figure 3—figure supplement 2E.
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| reg | 1.60E-03 | 2 | 7.99E-04 | 0.681 | 0.512 |
| day | 0.017 | 2 | 8.71E-03 | 7.426 | 1.73E-03 |
| reg:day | 2.53E-03 | 4 | 6.31E-04 | 0.538 | 0.708 |
| Error | 0.049 | 42 | 1.17E-03 | ||
| Total | 0.070 | 50 |
Post hoc comparison of median within-function group confusion across imaging day and region Figure 3—figure supplement 2E.
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D2 OT,d1 | D2 OT,d3 | –0.055 | 0.009 | 0.074 | 1.000 |
| D2 OT,d1 | D2 OT,d6 | –0.033 | 0.031 | 0.096 | 0.804 |
| D2 OT,d3 | D2 OT,d6 | –0.042 | 0.022 | 0.087 | 0.967 |
| D1 OT,d1 | D1 OT,d3 | –0.063 | 0.002 | 0.066 | 1 |
| D1 OT,d1 | D1 OT,d6 | –0.034 | 0.031 | 0.095 | 0.828 |
| D1 OT,d3 | D1 OT,d6 | –0.036 | 0.029 | 0.093 | 0.871 |
| VP,d1 | VP,d3 | –0.060 | 0.011 | 0.082 | 1 |
| VP,d1 | VP,d6 | –0.005 | 0.066 | 0.136 | 0.089 |
| VP,d3 | VP,d6 | –0.016 | 0.054 | 0.125 | 0.255 |
| D2 OT,d1 | D1 OT,d1 | –0.061 | 0.004 | 0.068 | 1 |
| D2 OT,d1 | VP,d1 | –0.067 | 0.001 | 0.069 | 1 |
| D1 OT,d1 | VP,d1 | –0.071 | –0.003 | 0.065 | 1 |
| D2 OT,d3 | D1 OT,d3 | –0.068 | –0.004 | 0.061 | 1 |
| D2 OT,d3 | VP,d3 | –0.065 | 0.003 | 0.070 | 1 |
| D1 OT,d3 | VP,d3 | –0.061 | 0.006 | 0.074 | 1 |
| D2 OT,d6 | D1 OT,d6 | –0.062 | 0.003 | 0.067 | 1 |
| D2 OT,d6 | VP,d6 | –0.033 | 0.035 | 0.103 | 0.756 |
| D1 OT,d6 | VP,d6 | –0.036 | 0.032 | 0.100 | 0.828 |
Two-way ANOVA for effect of imaging day and region on the mean accuracy for linear classification of {S vs. X} using population data (Figure 5—figure supplement 1B).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.010 | 2 | 4.92E-03 | 0.868 | 0.427 |
| day | 0.178 | 2 | 0.089 | 15.734 | 7.95E-06 |
| region:day | 0.064 | 4 | 0.016 | 2.846 | 3.56E-02 |
| Error | 0.238 | 42 | 5.67E-03 | ||
| Total | 0.476 | 50 |
Post hoc comparison of mean {S vs. X} accuracy across imaging day and region (Figure 5—figure supplement 1B).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D1 OT,d1 | D2 OT,d1 | –0.108 | 0.034 | 0.176 | 0.997 |
| D1 OT,d1 | VP,d1 | –0.018 | 0.131 | 0.280 | 0.127 |
| D2 OT,d1 | VP,d1 | –0.052 | 0.097 | 0.246 | 0.476 |
| D1 OT,d3 | D2 OT,d3 | –0.081 | 0.061 | 0.203 | 0.889 |
| D1 OT,d3 | VP,d3 | –0.153 | –0.004 | 0.145 | 1 |
| D2 OT,d3 | VP,d3 | –0.214 | –0.065 | 0.084 | 0.880 |
| D1 OT,d6 | D2 OT,d6 | –0.142 | 0.000 | 0.142 | 1 |
| D1 OT,d6 | VP,d6 | –0.202 | –0.053 | 0.096 | 0.960 |
| D2 OT,d6 | VP,d6 | –0.202 | –0.053 | 0.096 | 0.960 |
| D1 OT,d1 | D1 OT,d3 | –0.190 | –0.048 | 0.094 | 0.971 |
| D1 OT,d1 | D1 OT,d6 | –0.214 | –0.072 | 0.070 | 0.765 |
| D1 OT,d3 | D1 OT,d6 | –0.166 | –0.024 | 0.118 | 1 |
| D2 OT,d1 | D2 OT,d3 | –0.163 | –0.021 | 0.121 | 1 |
| D2 OT,d1 | D2 OT,d6 | –0.248 | –0.106 | 0.036 | 0.288 |
| D2 OT,d3 | D2 OT,d6 | –0.227 | –0.085 | 0.057 | 0.575 |
| VP,d1 | VP,d3 | –0.338 | –0.183 | –0.027 | 1.12E-02 |
| VP,d1 | VP,d6 | –0.411 | –0.256 | –0.100 | 1.02E-04 |
| VP,d3 | VP,d6 | –0.229 | –0.073 | 0.082 | 0.830 |
Two-way ANOVA for effect of imaging day and region on the mean accuracy for linear classification of {S vs. P} using population data (Fig5-1C).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.020 | 2 | 0.010 | 1.776 | 0.182 |
| day | 0.255 | 2 | 0.128 | 22.389 | 2.41E-07 |
| region:day | 0.042 | 4 | 0.010 | 1.822 | 0.143 |
| Error | 0.240 | 42 | 5.71E-03 | ||
| Total | 0.543 | 50 |
Post hoc comparison of mean {S vs. P} accuracy across imaging day and region (Figure 5—figure supplement 1C).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D1 OT,d1 | D2 OT,d1 | –0.104 | 0.038 | 0.181 | 0.993 |
| D1 OT,d1 | VP,d1 | –0.051 | 0.098 | 0.248 | 0.453 |
| D2 OT,d1 | VP,d1 | –0.089 | 0.060 | 0.210 | 0.920 |
| D1 OT,d3 | D2 OT,d3 | –0.057 | 0.085 | 0.228 | 0.579 |
| D1 OT,d3 | VP,d3 | –0.134 | 0.015 | 0.165 | 1 |
| D2 OT,d3 | VP,d3 | –0.219 | –0.070 | 0.079 | 0.835 |
| D1 OT,d6 | D2 OT,d6 | –0.124 | 0.019 | 0.161 | 1 |
| D1 OT,d6 | VP,d6 | –0.192 | –0.043 | 0.107 | 0.989 |
| D2 OT,d6 | VP,d6 | –0.211 | –0.062 | 0.088 | 0.910 |
| D1 OT,d1 | D1 OT,d3 | –0.233 | –0.090 | 0.052 | 0.506 |
| D1 OT,d1 | D1 OT,d6 | –0.262 | –0.119 | 0.023 | 0.165 |
| D1 OT,d3 | D1 OT,d6 | –0.172 | –0.029 | 0.113 | 0.999 |
| D2 OT,d1 | D2 OT,d3 | –0.186 | –0.043 | 0.099 | 0.985 |
| D2 OT,d1 | D2 OT,d6 | –0.281 | –0.139 | 0.004 | 0.061 |
| D2 OT,d3 | D2 OT,d6 | –0.238 | –0.096 | 0.047 | 0.426 |
| VP,d1 | VP,d3 | –0.329 | –0.173 | –0.017 | 1.97E-02 |
| VP,d1 | VP,d6 | –0.417 | –0.261 | –0.105 | 7.71E-05 |
| VP,d3 | VP,d6 | –0.244 | –0.088 | 0.069 | 0.662 |
Two-way ANOVA for effect of imaging day and region on the accuracy for linear classification of {SK vs. ST} using population data (Figure 5—figure supplement 1D).
| Source | Sum Sq. | d.f. | Mean Sq. | F | Prob >F |
|---|---|---|---|---|---|
| region | 0.085 | 2 | 0.042 | 3.600 | 0.036 |
| day | 0.033 | 2 | 0.017 | 1.415 | 0.254 |
| region:day | 0.042 | 4 | 0.010 | 0.890 | 0.478 |
| Error | 0.493 | 42 | 0.012 | ||
| Total | 0.655 | 50 |
Post hoc comparison of {SK vs. ST} accuracy across imaging day and region (Figure 5—figure supplement 1D).
| Group A | Group B | Lower Limit | A-B | Upper Limit | p-value |
|---|---|---|---|---|---|
| D1 OT,d1 | D2 OT,d1 | –0.202 | 0.003 | 0.207 | 1 |
| D1 OT,d1 | VP,d1 | –0.121 | 0.094 | 0.308 | 0.879 |
| D2 OT,d1 | VP,d1 | –0.123 | 0.091 | 0.306 | 0.896 |
| D1 OT,d3 | D2 OT,d3 | –0.238 | –0.033 | 0.171 | 1 |
| D1 OT,d3 | VP,d3 | –0.216 | –0.002 | 0.213 | 1 |
| D2 OT,d3 | VP,d3 | –0.183 | 0.032 | 0.246 | 1 |
| D1 OT,d6 | D2 OT,d6 | –0.307 | –0.103 | 0.102 | 0.776 |
| D1 OT,d6 | VP,d6 | –0.135 | 0.079 | 0.294 | 0.950 |
| D2 OT,d6 | VP,d6 | –0.032 | 0.182 | 0.397 | 0.153 |
| D1 OT,d1 | D1 OT,d3 | –0.199 | 0.006 | 0.210 | 1 |
| D1 OT,d1 | D1 OT,d6 | –0.227 | –0.022 | 0.182 | 1 |
| D1 OT,d3 | D1 OT,d6 | –0.232 | –0.028 | 0.177 | 1 |
| D2 OT,d1 | D2 OT,d3 | –0.235 | –0.031 | 0.174 | 1 |
| D2 OT,d1 | D2 OT,d6 | –0.332 | –0.128 | 0.077 | 0.525 |
| D2 OT,d3 | D2 OT,d6 | –0.302 | –0.097 | 0.107 | 0.823 |
| VP,d1 | VP,d3 | –0.314 | –0.090 | 0.134 | 0.922 |
| VP,d1 | VP,d6 | –0.261 | –0.037 | 0.187 | 1 |
| VP,d3 | VP,d6 | –0.171 | 0.053 | 0.277 | 0.997 |





