DBT is a metabolic switch for maintenance of proteostasis under proteasomal impairment
Figures
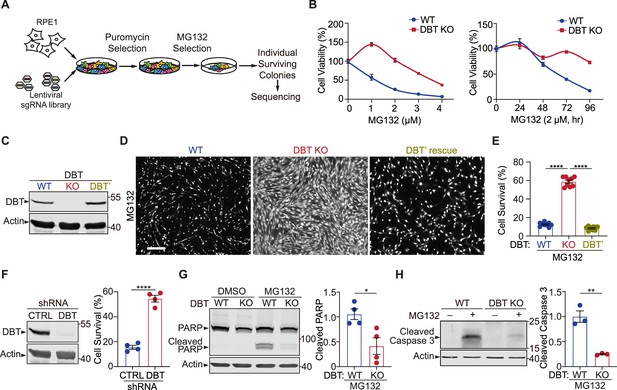
Genome-wide screen reveals that loss of dihydrolipoamide branched chain transacylase E2 (DBT) protects cells against the toxicity of proteasomal inhibition.
(A) Workflow of the CRISPR screen in retinal pigment epithelium (RPE1) cells, which were transduced with a lentiviral Genome-Scale CRISPR Knock-out (GeCKO) single guide RNA (sgRNA) library and selected for the sgRNA expression and then survival after treatments with the proteasome inhibitor MG132. Individual surviving cell colonies were collected for sequencing and subsequent analysis. (B) Left: The cytotoxicity analysis of wild-type (WT) and DBT knockout (KO) RPE1 cells treated with MG132 at different doses for 96 hr (n=3). Right: The time course analysis of MG132-induced cytotoxicity in the WT and DBT KO cells (n=3). (C) Immunoblot analysis of WT RPE1, DBT KO, and DBT’ cells. The DBT’ cells expressed an engineered DBT cDNA that resisted DBT-targeted Cas9 cleavage and rescued the DBT expression in the KO cells. (D) Cell viability was measured by Calcein-AM staining in WT RPE1, DBT KO, and DBT’ cells treated with MG132 (2 μM, 96 hr). Scale bar, 100 μm. (E) Quantification of the cell viability measured by Calcein-AM staining in (D) (n=9). (F) Left: Immunoblot analysis of RPE1 cells transfected with DBT shRNAs and non-targeting control shRNAs. Right: Quantification of the cell viability under treatment with MG132 (2 μM, 48 hr), as measured by Calcein-AM staining (n=4). (G) Immunoblotting and quantification of cleaved PARP as an MG132-induced cell death marker (n=4). (H) Immunoblotting and quantification of cleaved Caspase 3 as an MG132-induced cell death marker (n=3). Error bars represent means ± SEM. *p≤0.05; **p≤0.01; ****p≤0.0001.
-
Figure 1—source data 1
Original and uncropped blots for Figure 1C.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig1-data1-v1.zip
-
Figure 1—source data 2
Original and uncropped blots for Figure 1F.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig1-data2-v1.zip
-
Figure 1—source data 3
Original and uncropped blots for Figure 1G.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig1-data3-v1.zip
-
Figure 1—source data 4
Original and uncropped blots for Figure 1H.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig1-data4-v1.zip
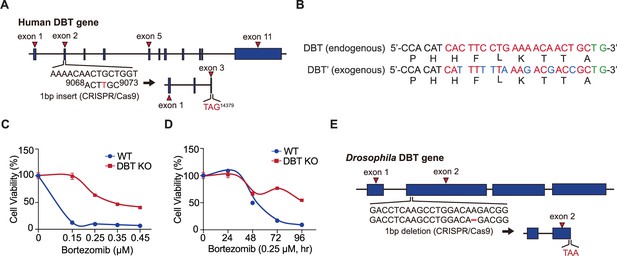
Schematics of CRISPR editing, dihydrolipoamide branched chain transacylase E2 (DBT) cDNA, and cell viability analysis.
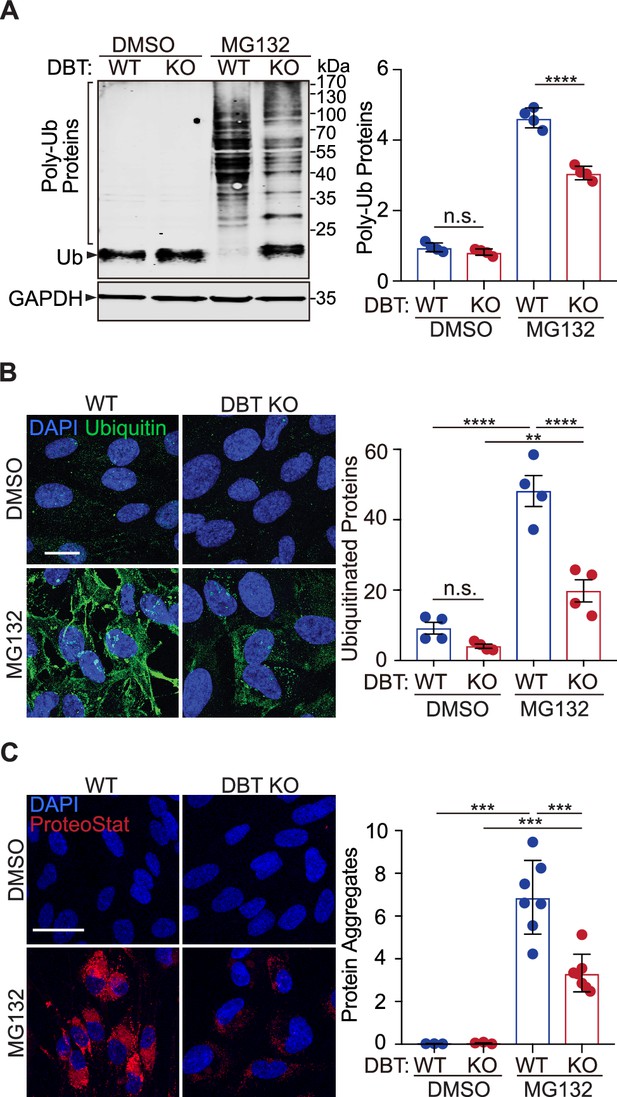
Loss of dihydrolipoamide branched chain transacylase E2 (DBT) decreased the accumulation of ubiquitinated proteins upon proteasomal inhibition.
(A) Wild-type (WT) and DBT knockout (KO) retinal pigment epithelium (RPE1) cells treated with MG132 (2 μM, 48 hr) or the DMSO solvent control were analyzed for accumulation of ubiquitinated proteins upon proteasomal inhibition with denaturing SDS-PAGE. The bar graph represents quantification of the high-molecular-weight poly-ubiquitinated proteins (n=4). (B) The cells treated with MG132 (2 μM, 72 hr) or the DMSO solvent control were analyzed for the levels of ubiquitinated protein with immunostaining. The bar graph represents the quantification of the anti-ubiquitin immunofluorescent signals (n=4 independent groups, each consisting of seven cells). Scale bar, 10 μm. (C) The cells treated with MG132 (2 μM, 72 hr) or the DMSO solvent control were stained with a dye that detects protein aggregates. The bar graph represents the quantification of the ProteoStat signals (n=7 biological replicates of DBT KO cells and three replicates of WT control cells). Scale bar, 20 μm. Error bars represent means ± SEM. ‘n.s.’, no significance; **p≤0.01; ***p≤0.001; ****p≤0.0001.
-
Figure 2—source data 1
Original and uncropped blots for Figure 2A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig2-data1-v1.zip
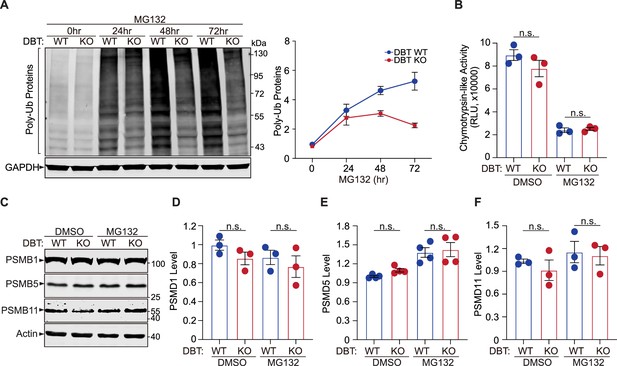
The enhanced clearance of poly-ubiquitinated proteins upon loss of dihydrolipoamide branched chain transacylase E2 (DBT) is not mediated by proteasomal degradation.
-
Figure 2—figure supplement 1—source data 1
Original and uncropped blots for Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Original and uncropped blots for Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig2-figsupp1-data2-v1.zip
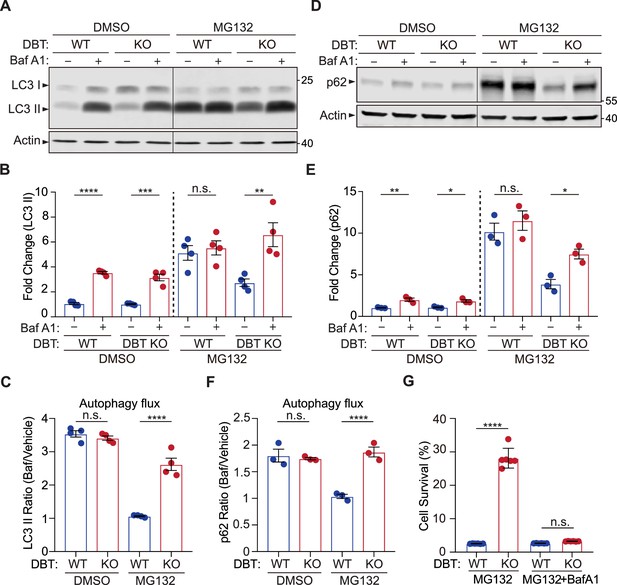
Loss of dihydrolipoamide branched chain transacylase E2 (DBT) preserves autophagic activities under proteasomal inhibition.
(A) Immunoblot analysis of LC3II levels in wild-type (WT) and DBT knockout (KO) retinal pigment epithelium (RPE1) cells treated with MG132 (2 μM, 48 hr) and with or without Baf A1 (Bafilomycin A1, 100 nM, 4 hr). (B) Quantification of the LC3II levels in (A) (n=4). (C) Quantification of the autophagic flux as measured by the ratios of LC3II levels before and after the Baf A1 treatment (n=4). (D) Immunoblot analysis of p62 in WT and DBT KO cells treated with MG132 (2 μM, 48 hr) with or without Baf A1 (Bafilomycin A1, 100 nM, 4 hr). (E) Quantification of the p62 levels in (D) (n=3). (F) Quantification of the autophagic flux as measured by the ratios of p62 levels before and after the Baf A1 treatment (n=3). (G) Cell viability analysis with crystal violet staining was performed on WT and DBT KO RPE1 cells treated with MG132 and with or without Baf A1 (n=6). Error bars represent means ± SEM. ‘n.s.’, no significance; *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001.
-
Figure 3—source data 1
Original and uncropped blots for Figure 3A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig3-data1-v1.zip
-
Figure 3—source data 2
Original and uncropped blots for Figure 3D.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig3-data2-v1.zip
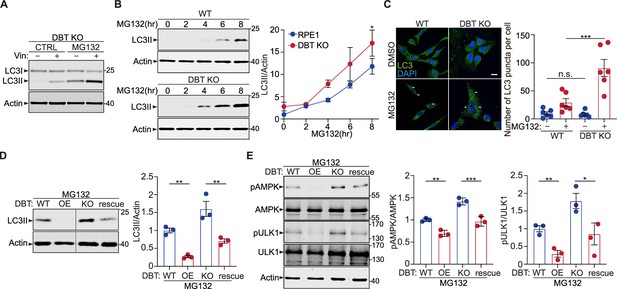
Loss of dihydrolipoamide branched chain transacylase E2 (DBT) promotes cellular resistance to MG132-induced toxicity through the AMP-activated protein kinase (AMPK) signaling pathway.
-
Figure 3—figure supplement 1—source data 1
Original and uncropped blots for Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Original and uncropped blots for Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig3-figsupp1-data2-v1.zip
-
Figure 3—figure supplement 1—source data 3
Original and uncropped blots for Figure 3—figure supplement 1D.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig3-figsupp1-data3-v1.zip
-
Figure 3—figure supplement 1—source data 4
Original and uncropped blots for Figure 3—figure supplement 1E.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig3-figsupp1-data4-v1.zip
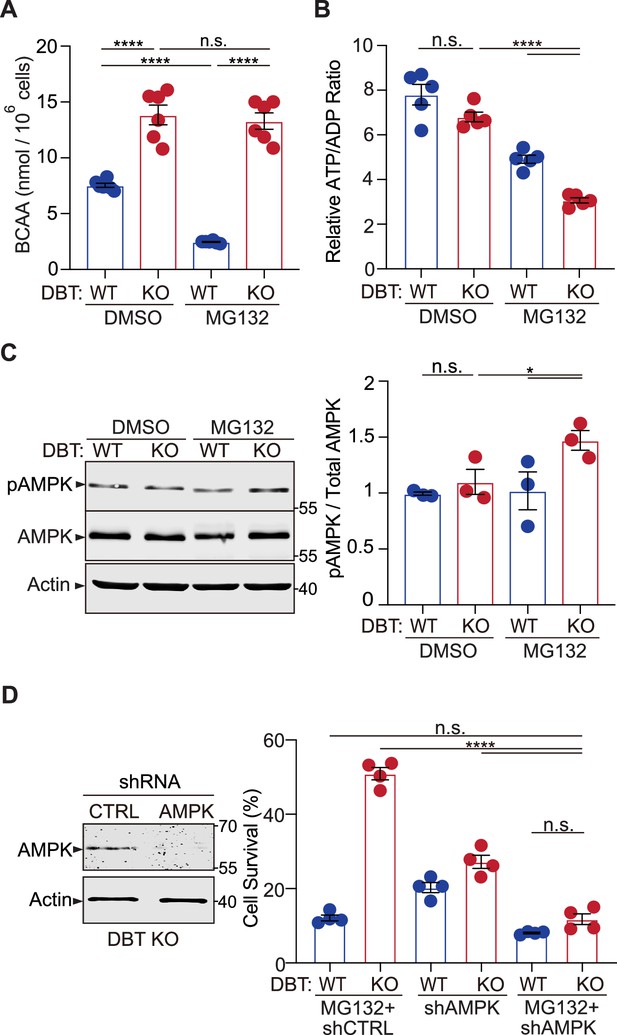
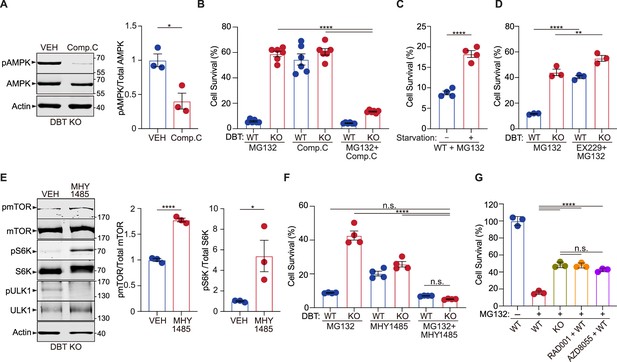
Loss of dihydrolipoamide branched chain transacylase E2 (DBT) activates AMP-activated protein kinase (AMPK) under proteasomal inhibition through energy regulation.
(A) Intracellular branched-chain amino acid (BCAA) levels were measured in wild-type (WT) and DBT knockout (KO) retinal pigment epithelium (RPE1) cells treated with MG132 (2 μM, 48 hr) (n=6). (B) Intracellular ATP/ADP ratios were measured in WT and DBT KO RPE1 cells treated with MG132 (2 μM, 48 hr) (n=5). (C) Activation of AMPK in DBT KO RPE1 cells treated with MG132 (2 μM, 48 hr), as indicated by the increase in the levels of phosphorylated AMPK (n=3). (D) The knockdown of AMPK by specific small hairpin RNAs (shRNAs) abolished the protective effects of loss of DBT against MG132-induced toxicity in DBT KO RPE1 cells, as indicated by the cell viability measured with crystal violet staining (n=4). Error bars represent means ± SEM. ‘n.s.’, no significance; *p≤0.05; ****p≤0.0001.
-
Figure 4—source data 1
Original and uncropped blots for Figure 4C.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig4-data1-v1.zip
-
Figure 4—source data 2
Original and uncropped blots for Figure 4D.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig4-data2-v1.zip
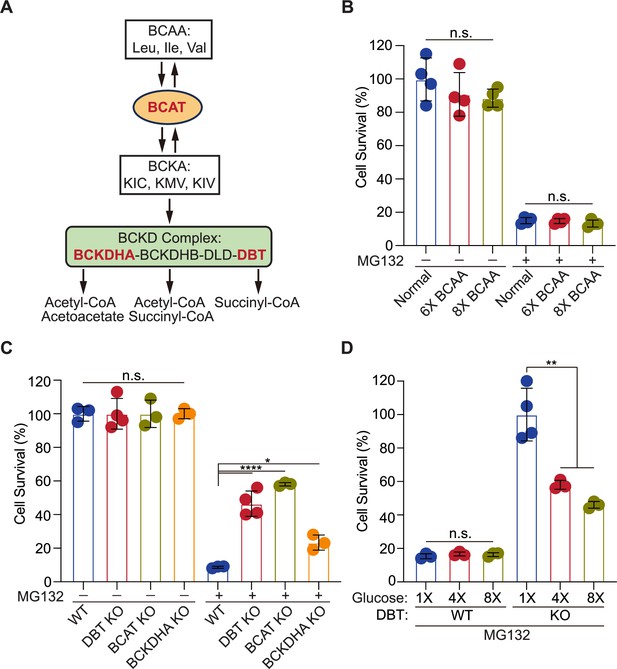
The energy depletion but not branched-chain amino acid (BCAA) accumulation mediates the resistance of dihydrolipoamide branched chain transacylase E2 (DBT) knockout cells to MG132-induced proteotoxicity.
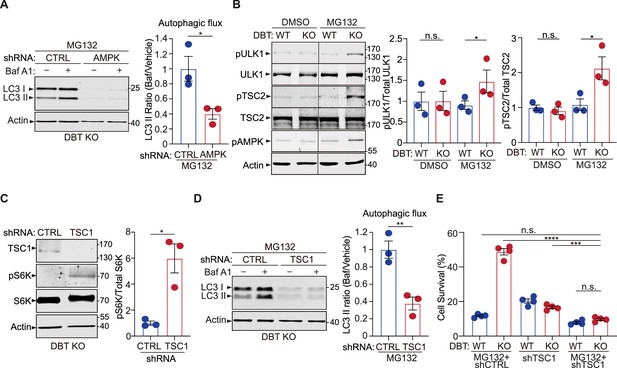
AMP-activated protein kinase (AMPK) downstream signaling enhances autophagy upon dihydrolipoamide branched chain transacylase E2 (DBT) deficiency and proteasomal inhibition.
(A) Immunoblot analysis of the autophagy marker LC3II in DBT knockout (KO) retinal pigment epithelium (RPE1) cells after the knockdown of AMPK using small hairpin RNAs (shRNAs) versus non-targeting control shRNAs (CTRL), under MG132 treatment conditions (2 μM, 48 hr), with or without the Baf A1 treatment. The autophagic flux is measured by calculating the ratios of LC3II protein levels with the Baf A1 treatment to those without the Baf A1 treatment (n=3). (B) Immunoblot analysis of the AMPK downstream effectors that regulate mTOR activities, including ULK1 and TSC2, in WT and DBT KO RPE1 cells with or without treatment with MG132 (2 μM, 48 hr). The activities of these regulators are quantified by measuring the levels of phosphorylation of ULK1-S371, TSC2-S1387, and AMPK-T172 (n=3). (C) Immunoblot analysis of the mTOR downstream marker S6K and its phosphorylation in DBT KO RPE1 cells after the knockdown of the negative regulator of mTOR, TSC1, using shRNAs versus control shRNAs (CTRL), under MG132 treatment conditions (2 μM, 48 hr) (n=3). (D) Immunoblot analysis of LC3II and quantification of the autophagic flux in DBT KO RPE1 cells after the knockdown of TSC1 under MG132 treatment conditions (2 μM, 48 hr) with or without the Baf A1 treatment (n=3). (E) Cell viability analysis with crystal violet staining of WT and DBT KO RPE1 cells after the knockdown of TSC1 using shRNAs versus control shRNAs (CTRL), under MG132 treatment conditions (2 μM, 48 hr) (n=4). Error bars represent means ± SEM. ‘n.s.’, no significance; *p≤0.05; **p≤0.01; ****p≤0.0001.
-
Figure 5—source data 1
Original and uncropped blots for Figure 5A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig5-data1-v1.zip
-
Figure 5—source data 2
Original and uncropped blots for Figure 5B.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig5-data2-v1.zip
-
Figure 5—source data 3
Original and uncropped blots for Figure 5C.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig5-data3-v1.zip
-
Figure 5—source data 4
Original and uncropped blots for Figure 5D.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig5-data4-v1.zip
Loss of dihydrolipoamide branched chain transacylase E2 (DBT) activates AMP-activated protein kinase (AMPK) and inhibits mTOR signaling.
-
Figure 5—figure supplement 1—source data 1
Original and uncropped blots for Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Original and uncropped blots for Figure 5—figure supplement 1E.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig5-figsupp1-data2-v1.zip
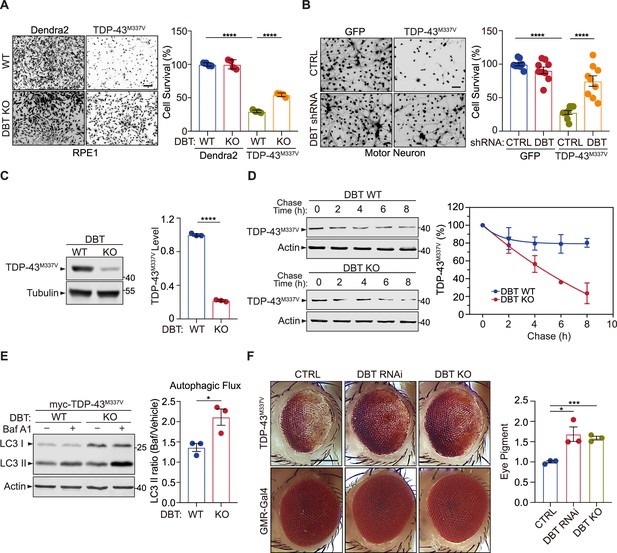
Loss of dihydrolipoamide branched chain transacylase E2 (DBT) protects against proteotoxicity of mutant TDP-43 in mammalian neurons and Drosophila models.
(A) Cell toxicity of amyotrophic lateral sclerosis (ALS)-linked TDP-43M337V expressed in wild-type (WT) and DBT knockout (KO) retinal pigment epithelium (RPE1) cells, with Dendra2 as a control, as measured by Calcein-AM staining (n=4). Scale bar, 300 μm. (B) Neuronal toxicity of TDP-43M337V expressed in mouse embryonic stem (ES) cell-differentiated motor neurons with or without DBT small hairpin RNA (shRNA)-mediated knockdown, compared to that of GFP control, as measured by Calcein-AM staining (n=9). Scale bar, 300 μm. (C) TDP-43M337V protein steady-state levels are significantly lower in DBT KO RPE1 cells than in WT control cells, as measured by immunoblot analysis (n=3). (D) The half-life of TDP-43M337V protein as measured in cycloheximide chase assays is significantly shorter in the DBT KO cells than in WT RPE1 control cells (n=3 independent experiments; p=0.0326). (E) Immunoblot analysis of the autophagy marker LC3II and quantification of the autophagic flux in WT and DBT KO RPE1 cells transfected with myc-TDP-43M337V with or without the Baf A1 treatment. The autophagic flux is measured by calculating the ratios of LC3II protein levels with the Baf A1 treatment to those without the Baf A1 treatment (n=3). (F) The reduction of DBT by RNAi or CRISPR led to strongly suppressed eye degeneration phenotypes in the TDP-43M337V fly strain when compared with the control Luc RNAi (CTRL). The eye degeneration phenotypes were quantified by measuring the pigment content in adult eyes (n=3 independent groups with each containing fly heads from four males and fpur females). Scale bar, 100 μm. Error bars represent means ± SEM. *p≤0.05; ***p≤0.001; ****p≤0.0001.
-
Figure 6—source data 1
Original and uncropped blots for Figure 6C.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig6-data1-v1.zip
-
Figure 6—source data 2
Original and uncropped blots for Figure 6D.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig6-data2-v1.zip
-
Figure 6—source data 3
Original and uncropped blots for Figure 6E.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig6-data3-v1.zip
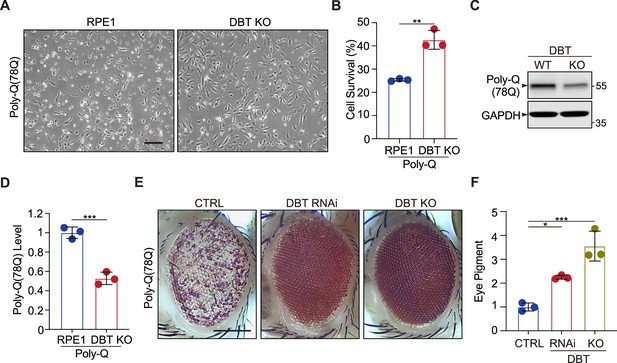
Loss of dihydrolipoamide branched chain transacylase E2 (DBT) protects against proteotoxicity of polyQ in mammalian neurons and Drosophila models.
-
Figure 6—figure supplement 1—source data 1
Original and uncropped blots for Figure 6—figure supplement 1C.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig6-figsupp1-data1-v1.zip
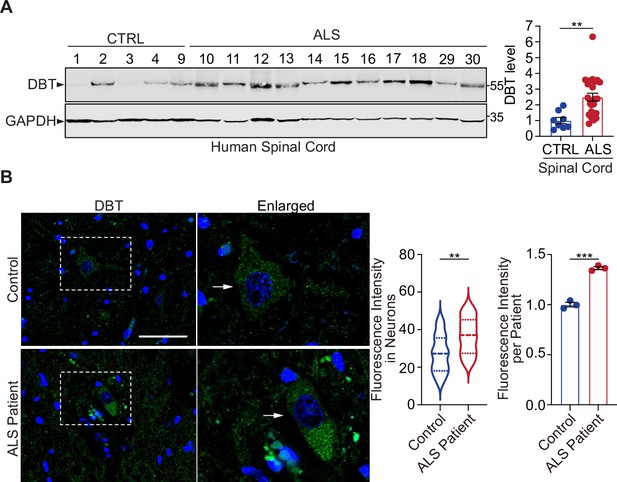
Dihydrolipoamide branched chain transacylase E2 (DBT) is abnormally upregulated in amyotrophic lateral sclerosis (ALS) patient neurons.
(A) Immunoblot analysis of human spinal cord tissues from ALS patients and non-neurological controls (n=24 ALS cases and eight non-neurological control cases). (B) Fluorescent immunostaining against DBT in the spinal cords from ALS patients and an age-matched control cases indicates that the accumulation of DBT in the patient’s neurons as identified by their morphological characteristics (n=23 neurons from three ALS cases and n=24 neurons from three control cases). Arrows point to representative motor neurons. Scale bar, 50 μm. Error bars represent means ± SEM. **p≤0.01.
-
Figure 7—source data 1
Original and uncropped blots for Figure 7A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig7-data1-v1.zip
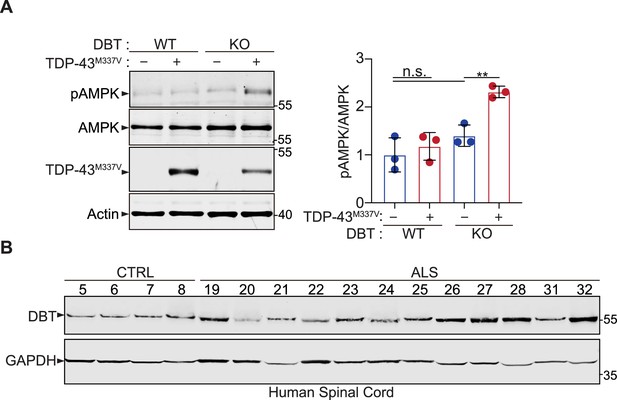
Analyses of the resistance of dihydrolipoamide branched chain transacylase E2 (DBT) knockout (KO) cells to TDP-43 toxicity and the increased DBT protein levels in amyotrophic lateral sclerosis (ALS) patients’ tissues.
-
Figure 7—figure supplement 1—source data 1
Original and uncropped blots for Figure 7—figure supplement 1A.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig7-figsupp1-data1-v1.zip
-
Figure 7—figure supplement 1—source data 2
Original and uncropped blots for Figure 7—figure supplement 1B.
- https://cdn.elifesciences.org/articles/91002/elife-91002-fig7-figsupp1-data2-v1.zip
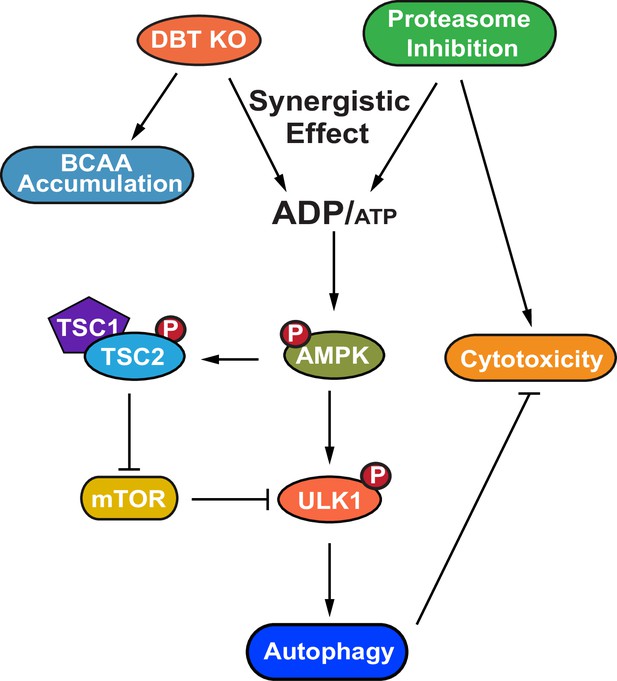
A model for the dihydrolipoamide branched chain transacylase E2 (DBT)-AMPK-autophagy signaling pathway.
A working model of the mechanism through which DBT acts as a metabolic switch for the maintenance of protein homeostasis through activation of autophagy under the condition of proteasomal inhibition. The loss of DBT leads to the accumulation of BCAAs as a result of the blocked catabolism of these amino acids, which tilts the balance of intracellular energy and reduces the levels of ATP, under the condition of proteasomal inhibition. This energy imbalance triggers the activation of AMPK, which then promotes autophagy through its regulation of mTOR and ULK1. BCAA, branched-chain amino acid; ATP, adenosine triphosphate; ADP, adenosine diphosphate; AMPK, AMP-activated protein kinase; TSC1, TSC complex subunit 1; TSC2, TSC complex subunit 2; ULK1, unc-51 like autophagy activating kinase 1.
Tables
The list of human patient’s tissues.
Table: List of patient’s tissues.
| Sample No. | Patient ID | Source | Clinical diagnosis | ALS pathology | Age of sampling | Gender | Region |
|---|---|---|---|---|---|---|---|
| 1 | 95 | TALS | CTRL | Non | 72 | M | SC-C |
| 2 | 103 | TALS | CTRL | Non | 22 | M | SC-C |
| 3 | 110 | TALS | CTRL | Non | 50 | M | SC-C |
| 4 | 108 | TALS | CTRL | Non | 72 | M | SC-C |
| 5 | 90015 | VABBB | CTRL | Non | 66 | M | SC-C |
| 6 | 90018 | VABBB | CTRL | Non | 82 | M | SC-C |
| 7 | 100012 | VABBB | CTRL | Non | 81 | F | SC-C |
| 8 | 120016 | VABBB | CTRL | Non | 63 | F | SC-C |
| 9 | AZ160030 | VABBB | ALS | Yes (TDP-43 pathology) | 65 | M | SC-C |
| 10 | AZ140006 | VABBB | ALS | Yes (TDP-43 pathology) | 74 | M | SC-C |
| 11 | 110011 | VABBB | ALS | Yes (TDP-43 pathology) | 83 | M | SC-C |
| 12 | 140008 | VABBB | ALS | Yes (TDP-43 pathology) | 75 | M | SC-C |
| 13 | AZ150001 | VABBB | ALS | Yes (TDP-43 pathology) | 65 | M | SC-C |
| 14 | AZ140021 | VABBB | fALS | Yes | 63 | M | SC-C |
| 15 | AZ150004 | VABBB | ALS | Yes (TDP-43 pathology) | 67 | M | SC-C |
| 16 | 130022 | VABBB | ALS | Yes (TDP-43 pathology) | 48 | M | SC-C |
| 17 | 130025 | VABBB | ALS | Yes (TDP-43 pathology) | 77 | M | SC-C |
| 18 | AZ140023 | VABBB | ALS | Yes (TDP-43 pathology) | 68 | M | SC-C |
| 19 | 130020 | VABBB | ALS | Yes (TDP-43 pathology) | 78 | M | SC-C |
| 20 | 130014 | VABBB | ALS | Yes (TDP-43 pathology) | 70 | M | SC-C |
| 21 | 100007 | VABBB | fALS | Yes (TDP-43 pathology) | 61 | M | SC-C |
| 22 | 100040 | VABBB | ALS | Yes | 88 | M | SC-C |
| 23 | 120015 | VABBB | ALS | Yes (TDP-43 pathology) | 58 | M | SC-C |
| 24 | 90003 | VABBB | fALS | Yes | 73 | M | SC-C |
| 25 | 90005 | VABBB | ALS | Yes (TDP-43 pathology) | 65 | M | SC-C |
| 26 | 90020 | VABBB | fALS | Yes | 49 | F | SC-C |
| 27 | 100002 | VABBB | ALS | Yes (TDP-43 pathology) | 63 | M | SC-C |
| 28 | AZ140017 | VABBB | ALS | Yes (TDP-43 pathology) | 66 | M | SC-C |
| 29 | 38 | TALS | sALS | Yes (C9orf72 HRE) | 34 | F | SC-C |
| 30 | 88 | TALS | sALS | Yes (C9orf72 HRE) | 59 | M | SC-C |
| 31 | 92 | TALS | fALS | Yes (C9orf72 HRE) | 72 | M | SC-C |
| 32 | MY9 | TALS | ALS | Yes (C9orf72 HRE) | 62 | F | SC-C |
-
Abbreviations: ALS (Amyotrophic Lateral Sclerosis), fALS (familial amyotrophic lateral sclerosis), sALS (sporadic amyotrophic lateral sclerosis), CTRL (Control), M (Male), F (Female), TALS (Target ALS Human Postmortem Tissue Core), VABBB (VA Biorepository Brain Bank), SC-C (Spinal Cord-Cervical), and HRE (hexanucleotide repeat expansion). The mean age of the controls is 63.5 years versus 69.4 years for the patients.





