The mTOR pathway genes MTOR, Rheb, Depdc5, Pten, and Tsc1 have convergent and divergent impacts on cortical neuron development and function
Figures

Expression of RhebY35L, MTORS2215Y, Depdc5KO, PtenKO, and Tsc1KO leads to varying magnitudes of neuronal enlargement and mispositioning in the cortex.
(a) Diagram of the PI3K-mTORC1 pathway. Activation of mTOR complex 1 (mTORC1) signaling is controlled by positive (blue) and negative (purple) regulators within the pathway. (b) Diagram of overall experimental timeline and approach. in utero electroporation (IUE) was performed at E15.5. A cohort of animals was used for patch clamp recording at P26-51 and another cohort was used for histology at P28-43. (c) Representative images of tdTomato+ cells (red) and p-S6 staining (green) in mouse medial prefrontal cortex (mPFC) at P28-43. (d) Quantification of p-S6 staining intensity (normalized to the mean control) in tdTomato+ neurons. (e) Quantification of tdTomato+ neuron soma size. (f) Representative images of tdTomato+ neuron (red) placement and distribution in coronal mPFC sections. Red square on the diagram denotes the imaged area for all groups. CC, corpus callosum. (g) Quantification of tdTomato+ neuron placement in layer 2/3. Left diagram depicts the approach for analysis: the total number of tdTomato+ neurons within layer 2/3 (white square) was counted and expressed as a % of total neurons in the imaged area. Right bar graphs show the quantification. (h) Quantification of tdTomato+ neuron distribution across cortical layers. Left diagram depicts the approach for analysis: the imaged area was divided into 10 equal-sized bins across the cortex, spanning the corpus callosum to the pial surface (white grids); the total number of tdTomato+ neurons within each bin was counted and expressed as a % of total neurons in the imaged area. Right bar graphs show the quantification. For graphs (d, e): n=4–8 mice per group, with 6–15 cells analyzed per animal. For graphs (g, h): n=3–7 mice per group, with 1 brain section analyzed per animal. Statistical comparisons were performed using (d, e) nested one-way ANOVA (fitted to a mixed-effects model) to account for correlated data within individual animals, (g) one-way ANOVA, or (h) two-way repeated measures ANOVA. Post-hoc analyses were performed using Holm-Šídák multiple comparison test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. All data are reported as the mean of all neurons or brain sections in each group ± SEM.
-
Figure 1—source data 1
Summary statistics for Figure 1.
- https://cdn.elifesciences.org/articles/91010/elife-91010-fig1-data1-v1.docx
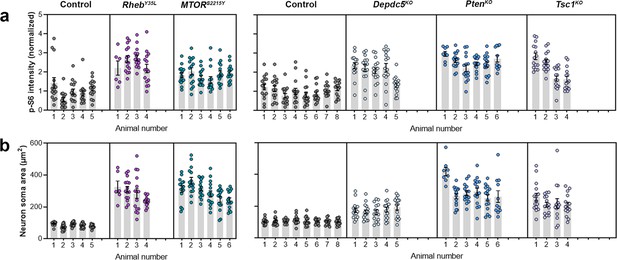
Distribution of p-S6 staining intensity and neuron soma size among individual animals at P28-43.
(a) Distribution of p-S6 staining intensity among individual animals. (b) Distribution of neuron soma size among individual animals.
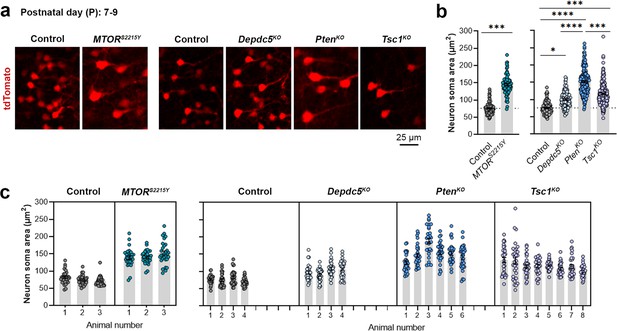
Neuron soma sizes at P7-9.
(a) Representative images of tdTomato+ cells (red) in mouse medial prefrontal cortex (mPFC) at P7-9. (b) Quantification of tdTomato+ neuron soma size. (c) Graph showing the distribution of neuron soma size among individual animals in each group. n=3–8 mice per group, with 30 cells analyzed per animal. Statistical comparisons were performed using a nested t-test (control vs. MTORS2215Y) or nested one-way ANOVA (control vs. Depdc5KO vs. PtenKO, vs. Tsc1KO) (fitted to a mixed-effects model) to account for correlated data within individual animals. Post-hoc analyses were performed using Holm-Šídák multiple comparison test. *p<0.05, ***p<0.001, ****p<0.0001. Data are reported as the mean of all neurons in each group ± SEM.
-
Figure 1—figure supplement 2—source data 1
Summary statistics for Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/91010/elife-91010-fig1-figsupp2-data1-v1.docx
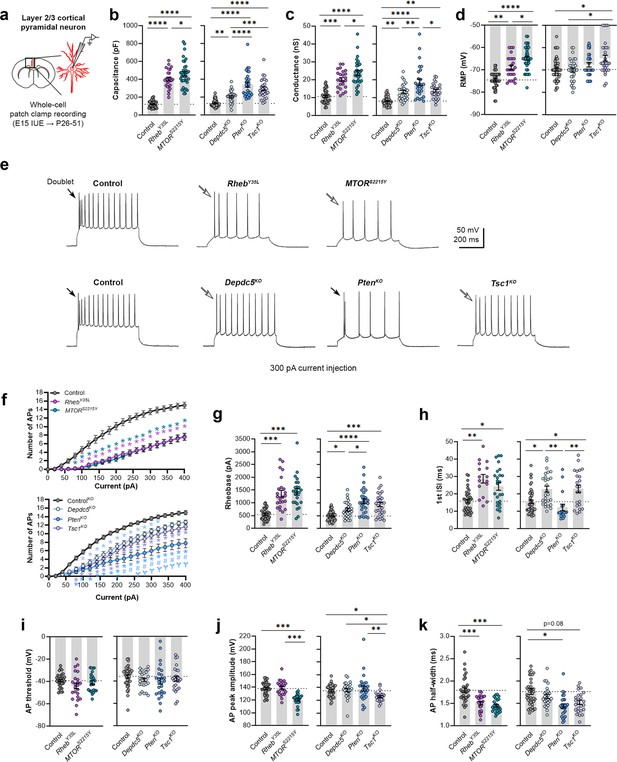
Expression of RhebY35L, MTORS2215Y, Depdc5KO, PtenKO, and Tsc1KO universally leads to decreased depolarization-induced excitability, but only RhebY35L, MTORS2215Y, and Tsc1KO expression leads to depolarized resting membrane potentials (RMPs).
(a) Diagram of experimental approach for whole-cell patch clamp recording. Recordings were performed in layer 2/3 pyramidal neurons in acute coronal slices from P26-51 mice, expressing either control, RhebY35L, MTORS2215Y, Depdc5KO, PtenKO, or Tsc1KO plasmids. (b–d) Graphs of membrane capacitance, resting membrane conductance, and RMP. (e) Representative traces of the action potential (AP) firing response to a 300 pA depolarizing current injection. (f) Input-output curves show the mean number of APs fired in response to 500 ms-long depolarizing current steps from 0 to 400 pA. Arrows point to initial spike doublets. (g–k) Graphs of rheobase, first ISI, AP threshold, AP peak amplitude, and AP half-width. For all graphs: n=5–10 mice per group, with 16–50 cells analyzed per animal. Statistical comparisons were performed using (b–d, g–k) nested one-way ANOVA (fitted to a mixed-effects model) to account for correlated data within individual animals or (f) mixed-effects ANOVA accounting for repeated measures. Post-hoc analyses were performed using Holm-Šídák multiple comparison test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, for the input-output curves in (f): *p<0.05 (vs. control), #<0.05 (vs. Depdc5KO), Ɏp<0.05 (vs. Tsc1KO). All data are reported as the mean of all neurons in each group ± SEM.
-
Figure 2—source data 1
Summary statistics for Figure 2.
- https://cdn.elifesciences.org/articles/91010/elife-91010-fig2-data1-v1.docx
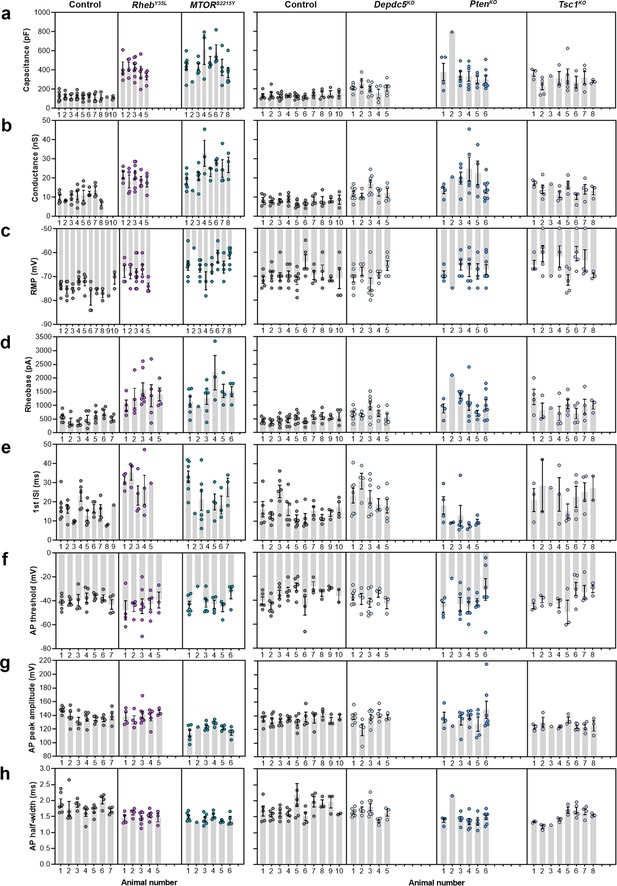
Distribution of membrane and action potential (AP) properties among individual animals at P26-51.
(a–c) Distribution of membrane capacitance, conductance, and resting membrane potential (RMP) among individual animals. (d-h) Distribution of rheobase, first ISI, AP threshold, AP peak amplitude, and AP half-width among individual animals.
Expression of RhebY35L, MTORS2215Y, Depdc5KO, PtenKO, and Tsc1KO leads to the abnormal presence of HCN4 channels with variations in functional expression.
(a) Representative images of tdTomato+ cells (red) and HCN4 staining (green) in mouse medial prefrontal cortex (mPFC) at P28-43. (b) Quantification of HCN4 intensity (normalized to the mean control) in tdTomato+ neurons. (c) Representative current traces in response to a series of 3 s-long hyperpolarizing voltage steps from –120 to –40 mV, with a holding potential of –70 mV. Current traces from the –40 and –50 mV steps were not included due to contamination from unclamped Na+ spikes. Orange lines denote the current traces at –90 mV. (d) IV curve obtained from Iss amplitudes. (e) ΔIV curve obtained from Ih amplitudes (i.e. ΔI, where ΔI=Iss – Iinst). (f) Graphs of Ih amplitudes at –90 mV. (g) Representative current traces in response to a series of 3 s long hyperpolarizing voltage steps from –110 mV to –50 mV in the MTORS2215Y condition pre- and post-zatebradine application. Orange lines denote the current traces at –90 mV. (h) IV curve obtained from Iss amplitudes in the MTORS2215Y condition pre- and post-zatebradine application. (i) ΔIV curve obtained from Ih amplitudes (i.e. ΔI) in the MTORS2215Y condition pre- and post-zatebradine application. Arrow points to the post-zatebradine Ih amplitude at –90 mV. (j) Representative traces of the zatebradine-sensitive current obtained after subtraction of the post- from the pre-zatebradine current traces in response to –110 mV to –50 mV voltage steps. Orange lines denote the current traces at –90 mV. (k) IV curve of the zatebradine-sensitive current obtained after subtraction of the post- from the pre-zatebradine current traces. (l) Graph of RMP in the control and MTORS2215Y conditions pre- and post-zatebradine application. Connecting lines denote paired values from the same neuron. For graph (b): n=4–8 mice per group, with 4–15 cells analyzed per animal. For graphs d, e, f: n=5–10 mice per group, with 24–47 cells analyzed per animal. For graphs h, i, k, l: n=4–6 neurons (paired). Statistical comparisons were performed using (b, f) nested ANOVA (fitted to a mixed-effects model) to account for correlated data within individual animals, (d, e) mixed-effects ANOVA accounting for repeated measures, or (h, i, l) two-way repeated measures ANOVA. Post-hoc analyses were performed using Holm-Šídák multiple comparison test. *p<0.05, **p<0.01, ****p<0.0001, for the IV curves in d, e, h, i: *p<0.05 (vs. control), #p<0.05 (vs. RhebY35L or Depdc5KO), Ɏp<0.05 (vs. PtenKO). All data are reported as the mean of all neurons in each group ± SEM.
-
Figure 3—source data 1
Summary statistics for Figure 3.
- https://cdn.elifesciences.org/articles/91010/elife-91010-fig3-data1-v1.docx
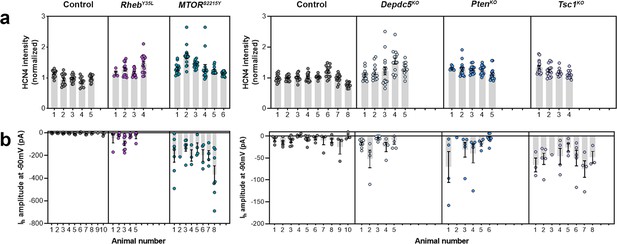
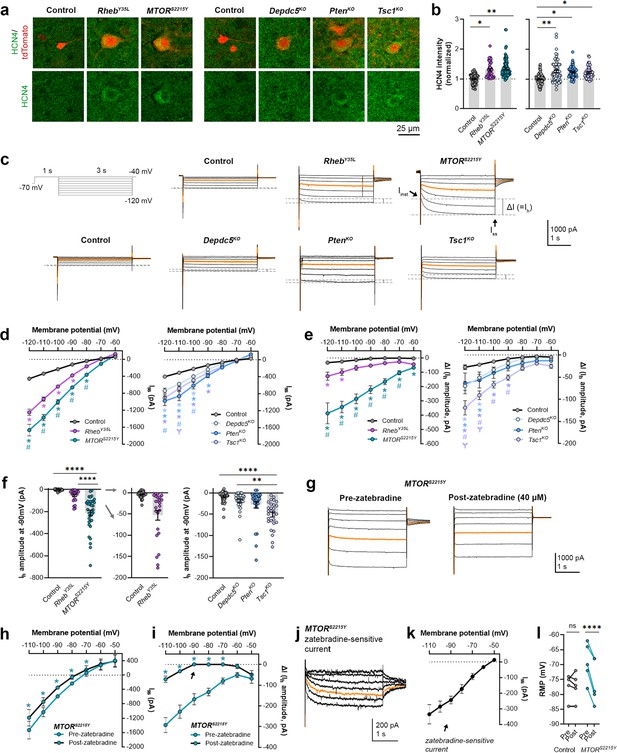
Distribution of HCN4 staining intensity and hyperpolarization-activated cation current (Ih) amplitudes (at –90 mV) among individual animals at P28-43 and P26-51, respectively.
(a) Distribution of HCN4 staining intensity among individual animals. (d-g) Distribution of Ih amplitudes (at –90 mV) among individual animals.
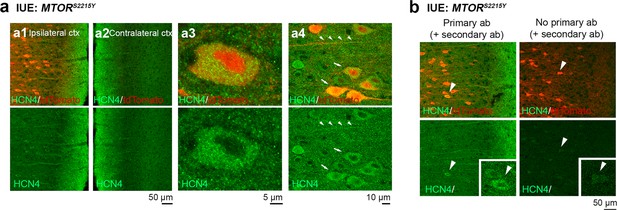
Additional images of HCN4 staining in medial prefrontal cortex (mPFC) sections from P28-43 mice expressing MTORS2215Y.
(a) Representative images of tdTomato+ cells (red) and HCN4 staining (green). Top panels show overlay images. Bottom panels show HCN4 single-channel images. (a1) and (a2) show the ipsilateral and contralateral cortex, respectively. (a3) and (a4) are high-magnification images demonstrating HCN4 staining within the cell. (b) Representative images of immunostaining with (left) or without (right) HCN4 primary antibodies (control for secondary antibody specificity).
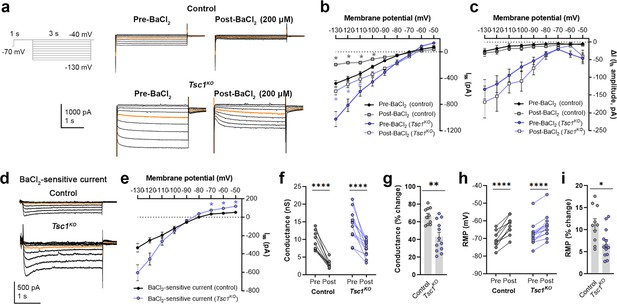
BaCl2 application decreases overall inward currents without affecting hyperpolarization-activated cation current (Ih) in Tsc1KO neurons.
(a) Representative current traces in response to a series of 3 s long hyperpolarizing voltage steps from –130 mV to –40 mV in the Tsc1KO condition pre- and post-BaCl2 application. Orange lines denote the current traces at –90 mV. (b) IV curve obtained from Iss amplitudes in the Tsc1KO condition pre- and post-BaCl2 application. (c) IV curve obtained from Ih amplitudes (i.e. ΔI, where ΔI=Iss – Iinst) in the Tsc1KO condition pre- and post-BaCl2 application. (d) Representative traces of the BaCl2-sensitive current obtained after subtraction of the post- from the pre-BaCl2 current traces. Orange lines denote the current traces at –90 mV. (e) IV curve of the BaCl2 -sensitive current obtained after subtraction of the post- from the pre-BaCl2 current traces. The isolated BaCl2-sensitive current reversed near –80 mV. (f) Graph of conductances (at –500 pA) in the control and Tsc1KO conditions pre- and post-BaCl2 application. Connecting lines denote paired values from the same neuron. (g) Graph of % change in conductances pre- and post-BaCl2 application in the control and Tsc1KO conditions. (h) Graph of resting membrane potential (RMP) in the control and Tsc1KO conditions pre- and post-BaCl2 application. Connecting lines denote paired values from the same neuron. (i) Graph of % change in RMP pre- and post-BaCl2 application in the control and Tsc1KO conditions. For all graphs: n=10–13 neurons per group. Statistical comparisons were performed using (b, c, e) mixed-effects ANOVA, (f, h) two-way repeated measures ANOVA, or (g, i) unpaired t-test. Post-hoc analyses were performed using Holm-Šídák multiple comparison test. *p<0.05, **p<0.01, ****p<0.0001. All data are reported as the mean of all neurons or brain sections in each group ± SEM.
-
Figure 3—figure supplement 3—source data 1
Summary statistics for Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/91010/elife-91010-fig3-figsupp3-data1-v1.docx
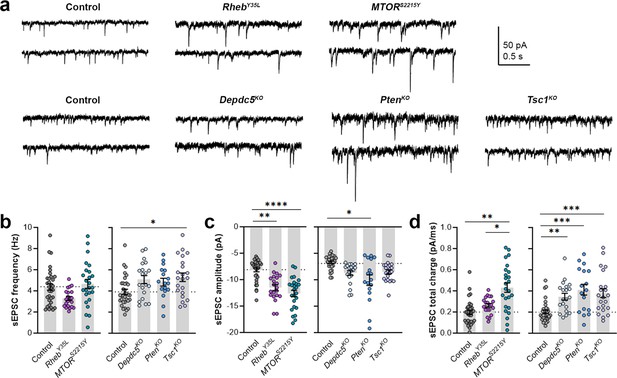
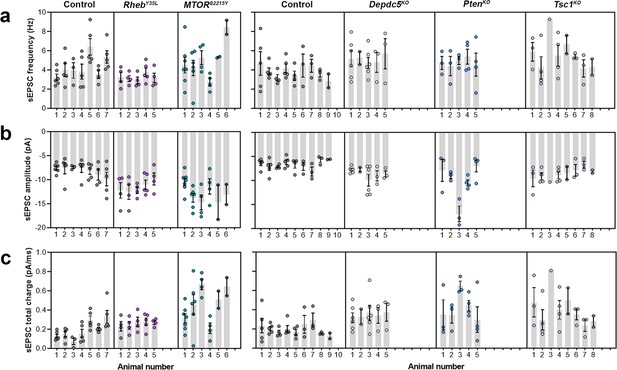
Expression of RhebY35L, MTORS2215Y, Depdc5KO, PtenKO, and Tsc1KO leads to different impacts on spontaneous excitatory postsynaptic current (sEPSC) properties.
(a) Representative sEPSC traces were recorded at a holding voltage of –70 mV. Top and bottom traces are from the same neuron. (b–d) Graphs of sEPSC frequency, amplitude, and total charge. For all graphs: n=5–9 mice per group, with 17–34 cells analyzed per animal. Statistical comparisons were performed using a nested ANOVA (fitted to a mixed effects model) to account for correlated data within individual animals. Post-hoc analyses were performed using Holm-Šídák multiple comparison test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. All data are reported as the mean of all neurons in each group ± SEM.
-
Figure 4—source data 1
Summary statistics for Figure 4.
- https://cdn.elifesciences.org/articles/91010/elife-91010-fig4-data1-v1.docx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | CD1, wildtype (time-pregnant females) | Charles River Laboratories | Strain code: 022, RRID:IMSR_CRL:022 | For in utero electroporation |
| Recombinant DNA reagent | pCAG-tdTomato (plasmid) | Addgene | Plasmid #83029, RRID:Addgene_83029 | Deposited by Dr. Angelique Bordey (previously generated by our lab) |
| Recombinant DNA reagent | pCAG-GFP (plasmid) | Addgene | Plasmid #11150, RRID:Addgene_11150 | Deposited by Dr. Connie Cepko |
| Recombinant DNA reagent | pCAG-tagBFP (plasmid) | Dr. Joshua Breunig, PMID:22776033 | - | Gift from Dr. Joshua Breunig. The generation of this plasmid has previously been described (Breunig et al., 2012). |
| Recombinant DNA reagent | pCAG-RhebY35L-T2A-tdTomato (plasmid) | This paper | - | The Rheb insert was synthesized and subcloned into a pCAG-Kir2.1Mut-T2A-tdTomato construct (Addgene plasmid #60644) via SmaI and BamHI sites, after excision of Kir2.1. Rheb insert sequence: NM_053075.3 (Mus musculus), mutation: Y35L |
| Recombinant DNA reagent | pCAG-MTORS2215Y (plasmid) | This paper | - | The MTOR insert was subcloned from a pcDNA3-FLAG-mTOR-SS2215Y construct (Addgene plasmid #69013) into a pCAG-tdTomato construct (Addgene plasmid #83029) via 5’AgeI and 3’NotI sites, after excision of tdTomato. MTOR insert sequence: NM_004958 (Homo sapiens), mutation: S2215Y |
| Recombinant DNA reagent | pX330-Control (pX330-empty) (plasmid) | Addgene | Plasmid #42230, RRID:Addgene_42230 | Deposited by Dr. Feng Zhang |
| Recombinant DNA reagent | pX330-Depdc5 (plasmid) | Dr. Stéphanie Baulac, PMID:29708508 | - | Gift from Dr. Stéphanie Baulac. The gRNA sequence (gRNA1) has previously been validated (Ribierre et al., 2018). gRNA sequence (5’ to 3’): GTCTGTGGTGATCACGC (17 nt) The gRNA targets exon 16 of Depdc5 (NM_001025426, Mus musculus), resulting in deleterious indels and inactivation of the gene |
| Recombinant DNA reagent | pX330-Pten (plasmid) | Addgene | Plasmid #59909, RRID:Addgene_59909 | Deposited by Dr. Tyler Jacks. The gRNA sequence (gRNA1) has previously been validated (Xue et al., 2014). gRNA sequence (5’ to 3’): AGATCGTTAGCAGAAACAAA (20 nt) The gRNA targets Pten (NM_008960, Mus musculus), resulting in deleterious indels and inactivation of the gene |
| Recombinant DNA reagent | pX330-Tsc1 (plasmid) | This paper | - | The gRNA sequence was synthesized based on previously published sequences (Lim et al., 2017) and subcloned into a pX330-empty vector. The specificity of the gRNA (gRNA T4) has previously been described and validated (Lim et al., 2017). gRNA sequence (5’ to 3’): CAGTGTGGAGGAGTCCAGCA (20 nt) The gRNA targets exon 3 of Tsc1 (NM_001289575, Mus musculus), resulting in deleterious indels and inactivation of the gene |
| Antibody | anti-p-S6 S240/244 (rabbit monoclonal) | Cell Signaling Technology | Cat # 5364, RRID:AB_10694233 | IF (1:1000) |
| Antibody | anti-HCN4 (rabbit polyclonal) | Alomone Labs | Cat # APC-052, RRID:AB_2039906 | IF (1:500) |
| Antibody | goat anti-rabbit IgG, Alexa Fluor Plus 647 | Invitrogen | Cat # A32733, RRID:AB_2633282 | IF (1:500) |
| Software, algorithm | Image J | NIH | RRID:SCR_003070 | - |
| Software, algorithm | Photoshop | Adobe | RRID:SCR_014199 | Version CC |
| Software, algorithm | pClamp | Molecular Devices | RRID:SCR_011323 | Version 10 |
| Software, algorithm | Prism | GraphPad Software | RRID:SCR_002798 | Version 9 |
Plasmid concentrations for control and experimental conditions.
| Group | Plasmid name | Plasmid concentration (µg/µl)* |
|---|---|---|
| Control (for RhebY35L littermates)† | pCAG-tdTomato | 3.5 |
| Control (for MTORS2215Y littermates)† | pCAG-GFP | 2.5 |
| pCAG-tdTomato | 1.0 | |
| RhebY35L | pCAG-RhebY35L-T2A-tdTomato | 2.5 |
| pCAG-GFP | 1.0 | |
| MTORS2215Y | pCAG-MTORS2215Y | 2.5 |
| pCAG-tdTomato | 1.0 | |
| Control | pX330-Control | 2.5 |
| pCAG-tdTomato | 1.0 | |
| pCAG-tagBFP‡ | 0.5 | |
| Depdc5KO | pX330-Depdc5 | 2.5 |
| pCAG-tdTomato | 1.0 | |
| pCAG-GFP‡ | 0.5 | |
| PtenKO | pX330-Pten | 2.5 |
| pCAG-tdTomato | 1.0 | |
| pCAG-GFP‡ | 0.5 | |
| Tsc1KO | pX330-Tsc1 | 2.5 |
| pCAG-tdTomato | 1.0 | |
| pCAG-GFP‡ | 0.5 |
-
*
Working plasmid solutions were diluted in water and contained 0.03% Fast Green dye to visualize the injection. For each embryo, 1.5 µl of the plasmid mixture was injected into the right ventricle.
-
†
Control groups for RhebY35L and MTORS2215Y contained pCAG tdTomato and pCAG-GFP +pCAG-tdTomato, respectively, to distinguish between control and experimental animals for each litter. The total plasmid concentration was kept equal between the two control groups. No differences were observed between the two groups, and therefore, they were combined into one control group.
-
‡
Equal concentrations of pCAG-tagBFP and pCAG-GFP were added into the control and experimental (Depdc5KO, PtenKO, and Tsc1KO) groups, respectively, to distinguish the control and experimental animals for each litter.