RAG1 and RAG2 non-core regions are implicated in leukemogenesis and off-target V(D)J recombination in BCR-ABL1-driven B-cell lineage lymphoblastic leukemia
Figures
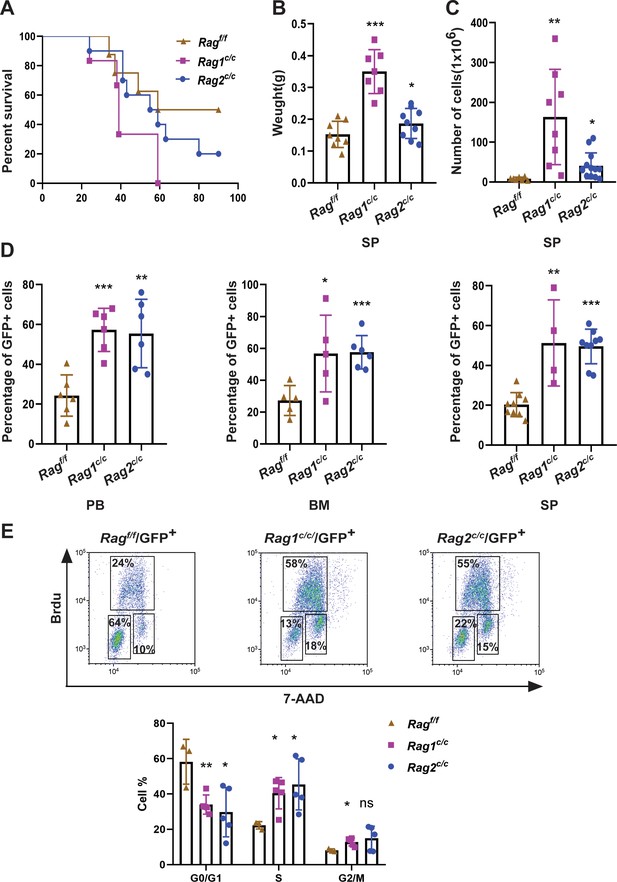
RAGc/cs give more aggressive leukemia in mice model of BCR-ABL1+ B-ALL.
(A) Kaplan–Meier survival curve for Ragf/f (n = 8), Rag1c/c (n = 6), and Rag2c/c (n = 10) recipient mice. The survival was calculated by Mantel–Cox test (Ragf/f vs Rag1c/c, p < 0.0381, Ragf/f vs Rag2c/c, p < 0.0412). (B) The spleen weights of Ragf/f, Rag1c/c, and Rag2c/c leukemic mice (Ragf/f, n = 8, Rag1c/c, n = 7, Rag2c/c, n = 9; Ragf/f vs Rag1c/c p < 0.0001, Ragf/f vs Rag2c/c, p = 0.1352). (C) The numbers of spleen cell in Ragf/f, Rag1c/c, and Rag2c/c leukemic mice (Ragf/f, n = 7, Rag1c/c, n = 8, Rag2c/c, n = 13; Ragf/f vs Rag1c/c, p = 0.0047, Ragf/f vs Rag2c/c, p = 0.0180). (D) The percentage of GFP+ cells in peripheral blood (PB) (Ragf/f, n = 6, Rag1c/c, n = 6, Rag2c/c, n = 6; Ragf/f vs Rag1c/c, p = 0.0003, Ragf/f vs Rag2c/c, p = 0.0035), bone marrow (BM, Ragf/f, n = 5, Rag1c/c, n = 5, Rag2c/c, n = 6; Ragf/f vs Rag1c/c, p = 0.0341, Ragf/f vs Rag2c/c, p = 0.0008), and spleen (SP, Ragf/f, n = 9, Rag1c/c, n = 4, cRAG2, n = 9; Ragf/f vs Rag1c/c, p = 0.0016, Ragf/f vs Rag2c/c, p < 0.0001) of Ragf/f, Rag1c/c, and Rag2c/c leukemic mice. (E) Representative flow cytometry plots of cell cycle arrest of leukemic cells in Ragf/f, Rag1c/c, and Rag2c/c mice. In the graph, the percentages of each phase of the cell cycle are summarized below (Ragf/f, n = 3, Rag1c/c, n = 5, Rag2c/c, n = 5; G0/G1, Ragf/f vs Rag1c/c, p = 0.0082, Ragf/fvs Rag2 c/c, p = 0.0279; S, Ragf/f vs Rag1 c/c, p = 0.0146, Ragf/f vs Rag2 c/c, p = 0.0370; G2/M, Ragf/f vs Rag1c/c, p = 0.0134, Ragf/f vs Rag2 c/c, p = 0.1507). In figures B, C, D, and J, error bars represent the mean ± standard deviation (s.d.), p values were calculated by Student’s t test and *p < 0.05, **p < 0.01, ***p < 0.001.
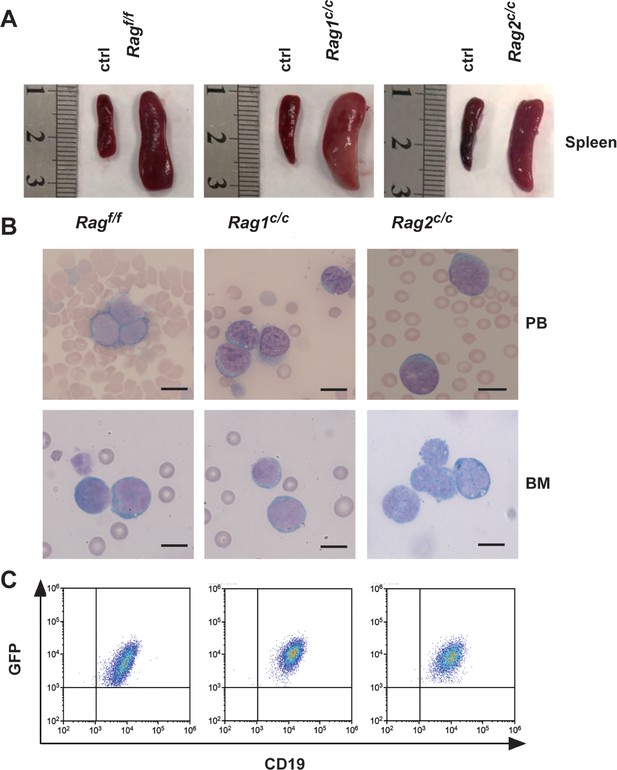
Construction of Ragf/f, Rag1c/c, and Rag2c/c, BCR-ABL1+ B-ALL mice models using bone marrow transplantation.
In the establishment of BCR-ABL1+ B-ALL mice models, Ragf/f, Rag1c/c, and Rag2c/c recipient mice after syngeneic lethal irradiation were transplanted with corresponding donor bone marrow cells transduced by MSCV-BCR-BAL1-IRES-GFP or MSCV-GFP retroviral supernatants. (A) Gross appearance of the spleen in Ragf/f, Rag1c/c, and Rag2c/c leukemic mice and corresponding control mice. (B) Peripheral blood (PB) and bone marrow (BM) lymphoblastic cells were stained by Wright–Giemsa. The scale bars represent 10 µm. (C) Bone marrow cells from Ragf/f, Rag1c/c, and Rag2c/c leukemic mice were examined by flow cytometry for the expression of green fluorescence protein (GFP) and CD19.
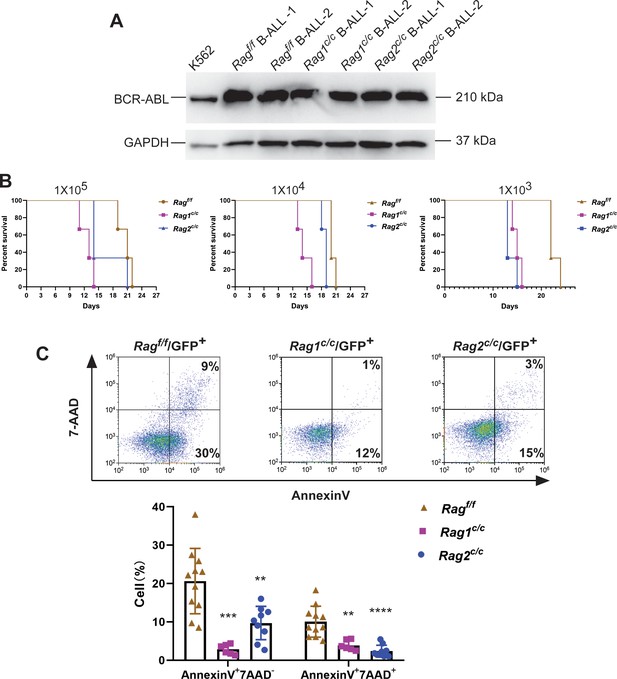
Biological behavior of leukemia in Ragf/f, Rag1c/c, and Rag2c/c BCR-ABL1+ B-ALL mouse model.
(A) BCR-ABL1 expressions in GFP+CD19+ leukemic cells were determined by western blotting. GAPDH protein was used as a loading control. The K562 and 293T cell lines served as the positive control and negative controls, respectively. (B) Survival of secondary transplant setting. Leukemia cells from primary recipients were recovered from the spleens and purified by GFP+ cell sorting. A total of 105,104, and 103 GFP+ leukemia cells originating from Ragf/f, Rag1c/c, or Rag2c/c BCR-ABL1+ B-ALL were transplanted into corresponding non-irradiated immunocompetent syngenetic recipient mice (RAG f/f, n = 3, Rag1c/c, n = 3, Rag2c/c, n = 3; survival days Rag f/f, 11–26 days, Rag1 c/c, 10–16 days, Rag2 c/c, 11–21 days in three different concentrations of GFP+ leukemic cells; Rag f/f vs Rag1 c/c, p = 0.0299, Ragf/f vs Rag2 c/c, p = 0.2286 in concentrations of 105, Ragf/f vs Rag1 c/c, p = 0.0246, Ragf/f vs Rag2c/c, p = 0.0295 in concentrations of 104, Ragf/f vs Rag1c/c, p = 0.0246, Ragf/f vs Rag2c/c, p = 0.0224 in concentrations of 103, by Mantel–Cox test). (C) Apoptosis was measured by flow cytometry (Annexin V and 7-AAD). The Annexin V+ and 7-AAD− cells were defined as early apoptotic cells, while Annexin V+ and 7-AAD+ cells were late apoptotic cells (Ragf/f, n = 11, Rag1c/c, n = 6, Rag2c/c, n = 9; early apoptotic cells: Ragf/f vs Rag1c/c, p = 0.0002, Ragf/f vs Rag2c/c, p = 0.0026; late apoptotic cells, Rag f/f vs Rag1 c/c, p = 0.0026, Rag f/f vs Rag2 c/c, p < 0.0001). Error bars represent the mean ± standard deviation (s.d.), p values were calculated by Student’s t test and **p < 0.01, ***p < 0.001, ****p < 0.0001.
-
Figure 1—figure supplement 2—source data 1
Original file for the western blot analysis in Figure 1—figure supplement 2A.
- https://cdn.elifesciences.org/articles/91030/elife-91030-fig1-figsupp2-data1-v1.pdf
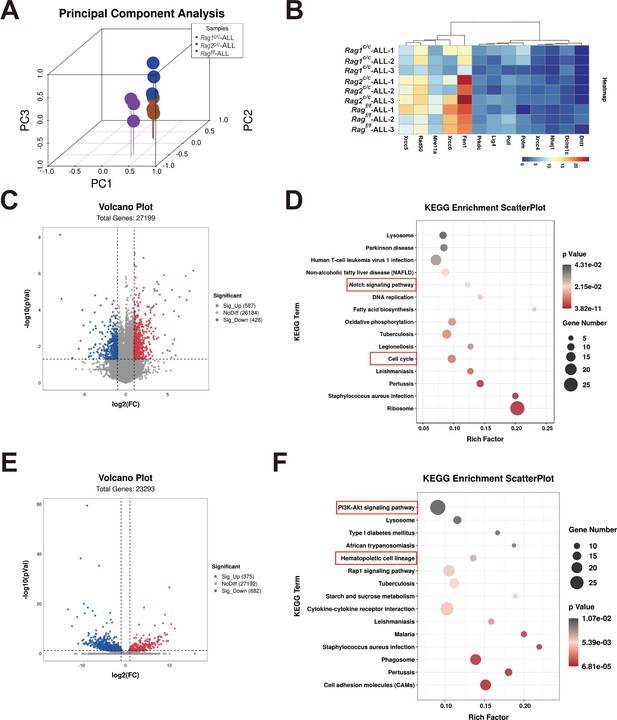
The genetic pathways in Ragf/f, Rag1C/C, and Rag2C/C BCR-ABL1+ lymphocytes.
mRNA sequence was performed in green fluorescence protein (GFP) and CD19 double positive cells. (A) Principal component analysis (PCA) showing the distribution of differentially expressed samples of Ragf/f, Rag1c/c, and Rag2c/c, BCR-ABL1+ B-ALL. (B) Heatmap of representative different expressed genes related to non-homologous end repair. The scale ranges from minimum (blue) to medium (yellow) to maximum (red) relative expression. (C) Volcano plot depicting log2 (fold change) (x-axis) and −log10 (p value) (y-axis) for differentially expressed genes (FC >2, p < 0.05) in GFP+CD19+ leukemic cells sorted from Ragf/f and Rag1c/c, BCR-ABL1+ B-ALL mice; upregulated (red) and downregulated (blue). n = 3 per group. (D) The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was conducted to identify the differentially expressed genes in Rag1c/c BCR-ABL1+ B-ALL. The top 15 pathways that exhibited significant differences were listed in this paragraph. The cell proliferation, apoptosis, and differentiation related pathway were highlighted in red squares. (E) Volcano plot depicting log2 (fold change) (x-axis) and −log10 (p value) (y-axis) for differentially expressed genes (FC >2, p < 0.05) in GFP+CD19+ leukemic cells sorted from Ragf/f and Rag2c/c, BCR-ABL1+ B-ALL mice; upregulated (red) and downregulated (blue). n = 3 per group. (F) KEGG analysis was conducted to identify the differentially expressed genes in Rag2c/c BCR-ABL1+ ALL. The top 15 pathways that exhibited significant differences were listed in this paragraph. The cell proliferation, apoptosis, and differentiation related pathway were highlighted in red squares.
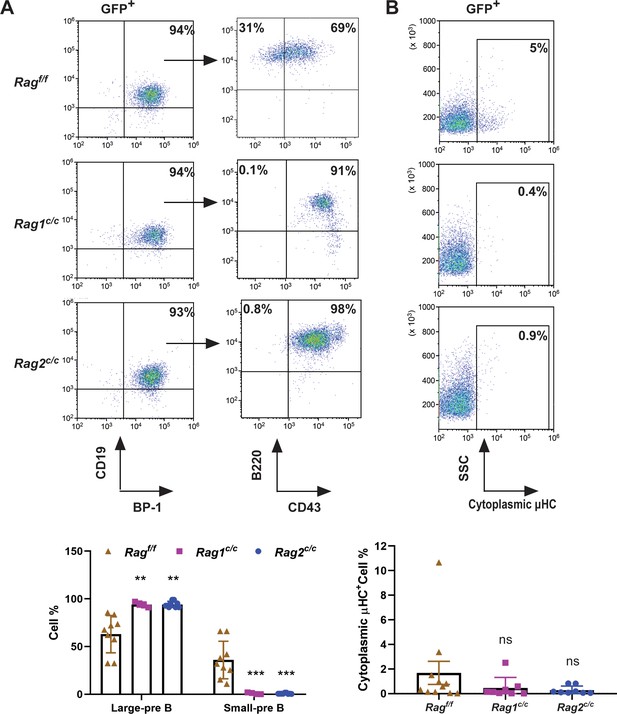
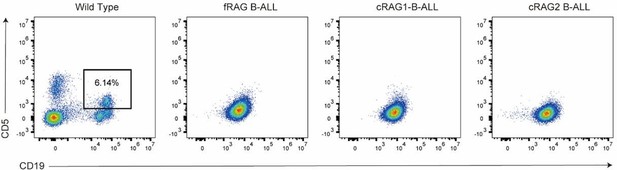
The non-core RAG region loss corresponds to a less mature cell surface phenotype.
(A) Flow cytometry analysis of the B-cell markers CD19, BP-1, B220, and CD43 on BCR-ABL1-transformed Ragf/f, Rag1c/c, and Rag2c/c leukemic bone marrow cells. The percentages of each phase of the B-cell stage are summarized in the bottom graph (Rag f/f, n = 9, Rag1 c/c, n = 4, Rag2 c/c, n = 9; Large-preB, Rag f/f, vs Rag1 c/c, p = 0.0349, Ragf/f, vs Rag2c/c, p = 0.0017; Small-pre-B, Rag f/f, vs Rag1c/c, p = 0.0141, Rag f/f, vs Rag2 c/c, p = 0.0005). The expression of the cytoplasmic μ chain was analyzed by flow cytometry. Representative samples are shown in (B), and the results from multiple samples analyzed in independent experiments are summarized in the bottom graph as the fraction of cells expressing cytoplasmic factors (Ragf/f, n = 11, Rag1c/c, n = 9, Rag2c/c, n = 8; Ragf/f vs Rag1c/c, p = 0.0312, Ragf/f, vs Rag2c/c, p = 0.0441). Error bars represent the mean ± standard deviation (s.d.), p values were calculated by Student’s t test and **p < 0.01, ***p < 0.001.
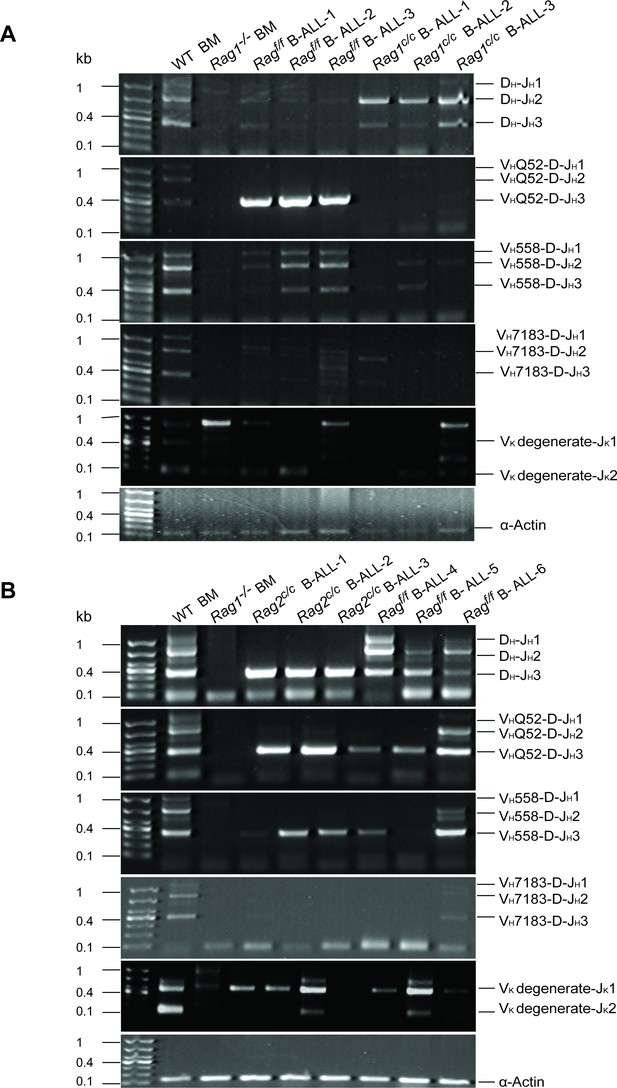
VDJ recombination in leukemic cells with different genetic backgrounds.
(A, B) VDJ recombination was analyzed by genomic PCR in GFP+CD19+ cell’s DNA from Ragf/f, Rag1c/c, or Rag2c/c leukemic cells. Genomic DNA from RAG1−/− bone marrow cells and WT spleen was used as negative and positive control, respectively (DNA Marker: 1000 bp, 700 bp, 500 bp, 400 bp, 300 bp, 200 bp, 100 bp).
-
Figure 2—figure supplement 1—source data 1
Original file for the PCR analysis in Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/91030/elife-91030-fig2-figsupp1-data1-v1.pdf
-
Figure 2—figure supplement 1—source data 2
Original file for the PCR analysis in Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/91030/elife-91030-fig2-figsupp1-data2-v1.pdf
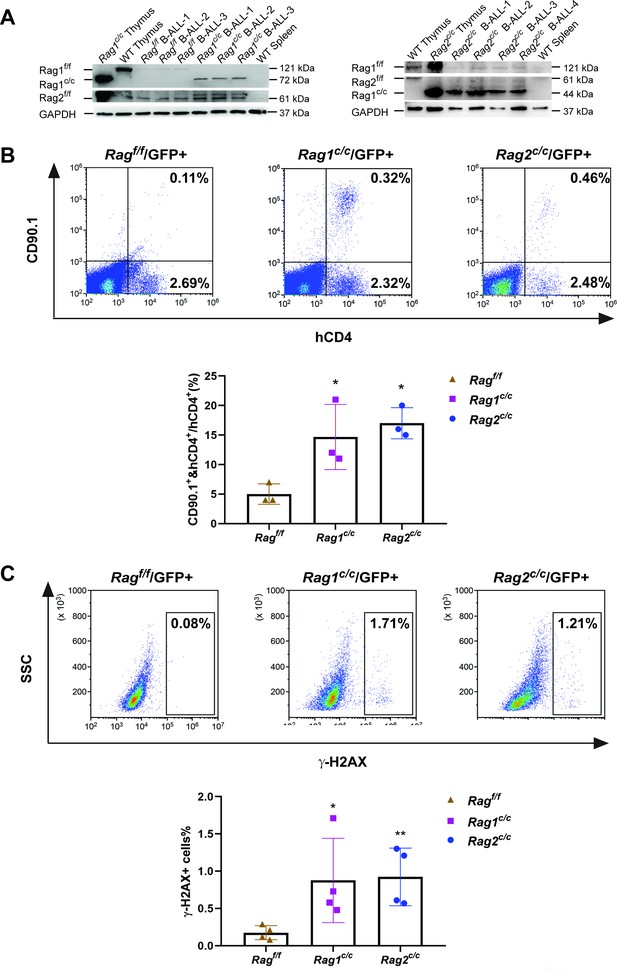
The non-core RAG region loss highlights genomic DNA damage.
(A) Western blotting analysis showed RAG1 and RAG2 expression in GFP+CD19+ leukemic cells originating from BCR-ABL1+ B-ALL in different genetic backgrounds. The experiment was repeated under the same conditions three times. (B) Rearrangement substrate retrovirus was transduced into leukemic cells. Flow cytometry was used to analyze the percentage of CD90.1- and hCD4-positive cells, and the percentage populations are shown in the bottom graph (Ragf/f, n = 3, Rag1c/c, n = 3, Rag2c/c, n = 3; Ragf/f, vs Rag1 c/c, p = 0.0002, Rag f/f, vs Rag2 c/c, p = 0.5865). (C) Flow cytometry analysis of ɣ-H2AX levels in Ragf/f, Rag1c/c, and Rag2c/c leukemic cells and the percentage of γ-H2AX-positive cell populations shown in the bottom graph (Rag f/f, n = 11, Rag1c/c, n = 8, Rag2c/c, n = 8; Ragf/f, vs Rag1c/c, p = 0.0505, Ragf/f, vs Rag2c/c, p = 0.0094). Error bars represent the mean ± standard deviation (s.d.), p values were calculated by Student’s t test and *p < 0.05, **p < 0.01.
-
Figure 3—source data 1
Original file for the western blot analysis in Figure 3A.
- https://cdn.elifesciences.org/articles/91030/elife-91030-fig3-data1-v1.pdf
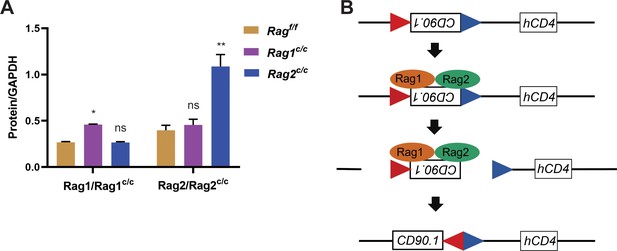
RAG protein expression levels and schematic diagram of the recombinant substrate vector.
(A) RAG1/cRAG1 and RAG2/cRAG2 protein levels were compared by western blot and ImageJ software in Ragf/f, Rag1c/c, and Rag2c/c, B-ALL cells. Error bars represent the mean ± standard deviation (s.d.). The p value was calculated by t test, **p < 0.01, *p < 0.05, ns p > 0.05. (B) The B-ALL cells were subjected to transformation with the recombinant substrate vector. In the event of expression of RAG recombinase in the leukemic cells, the recombination signal sequences (RSSs) flanking CD90.1 would be cleaved by RAG, thereby facilitating the positioning and expression of both CD90.1 and hCD4. In the absence of RAG expression, only hCD4 would be expressed.
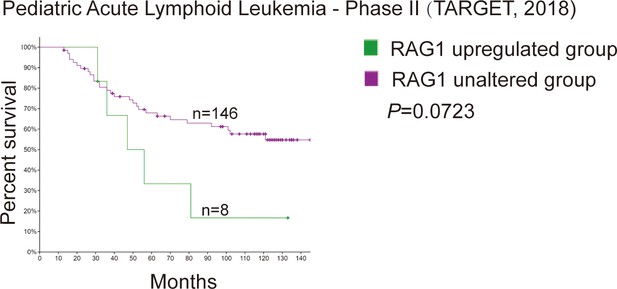
The relationship of RAG1 mRNA levels and survival of pediatric acute lymphoid leukemia.
The relationship of RAG1 mRNA levels and survival of pediatric acute lymphoid leukemia was research by cBioPortal (https://www.cbioportal.org/). RAG1 mRNA levels were studied in pediatric patients with ALL at the time of diagnosis. The patients were separated into two groups based on mRNA levels of RAG1 (mRNA expression z score relative to diploid sample, RNA sequence RPKM, RAG1 upregulated group, n = 8; RAG1 unaltered group, n = 146). The p values were calculated from the log-rank test, p = 0.0732.
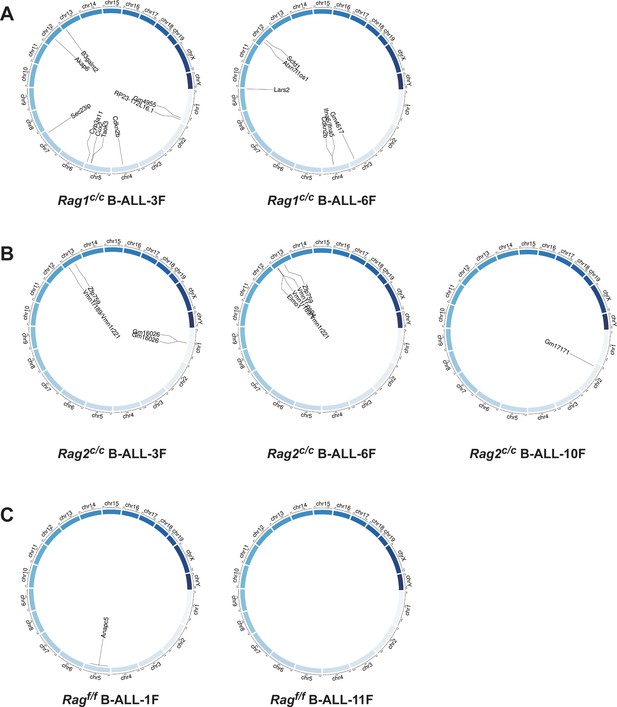
Structural alterations in BCR-ABL1+ B lymphocytes.
(A–C) Circos plot representation of all off-target recombination detected in the genome-wide analyses of Ragf/f, Rag1c/c, and Rag2c/c leukemic cells.
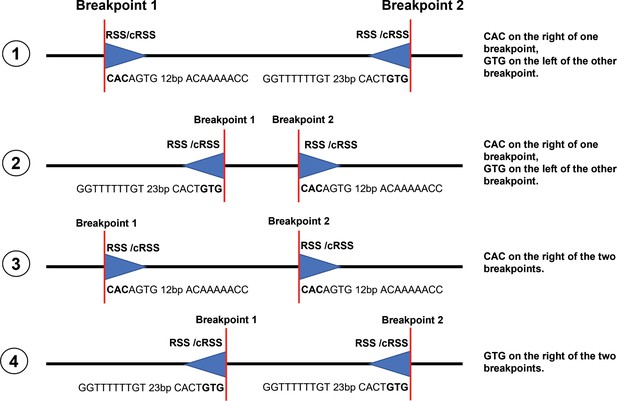
The criteria for identifying off-target recombination.
We adopted the criteria used in previous studies. First, a CAC must exist to the right (or GTG to the left) of both breakpoints, which includes the four RAG-mediated DNA fragmentation cases mentioned above, and second, it must occur within a specified distance from the breakpoint and the CAC distance-to-breakpoint value was set at 21 bp.
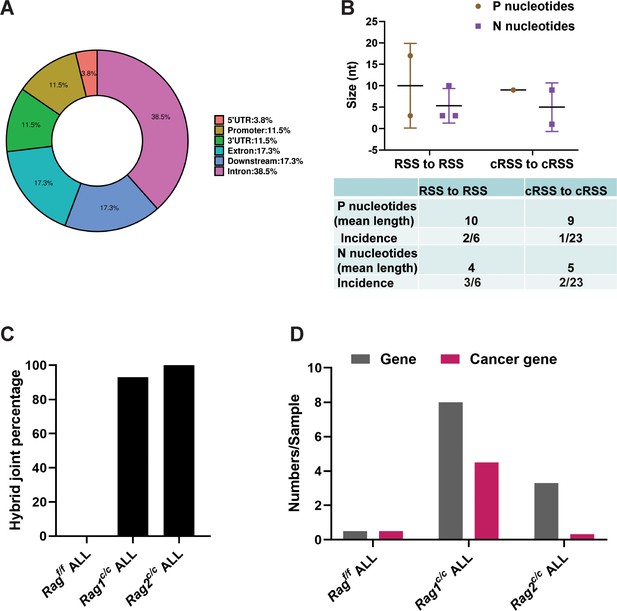
Overview and characteristics of off-target recombination in BCR-ABL1+ B-ALL leukemic cells from Ragf/f and Rag c/c mice.
(A) Exon–intron distribution profiles of 41 breakpoints generated by 24 structural variations (SVs). Gene body includes exon (n = 9; 17.3%) and intron (n = 20; 38.5%). Flanking sequence includes 3′UTR (n = 6; 11.5%), 5′UTR (n = 2; 3.8%), promoter (n = 6; 11.5%), and downstream (n = 9; 17.3%). (B) The off-target recombination was filtered and verified by whole genomic sequence and PCR, respectively. P and N nucleotides of recombination signal sequence (RSS) to RSS and cryptic RSS (cRSS) to cRSS were calculated in BCR-ABL1+ B-ALL. (C) Hybrid joint percentage generated by either Ragf/f, Rag1c/c, or Rag2c/c in BCR-ABL1+ B-ALL. It was 0, 100%, and 93% in Ragf/f, Rag1c/c, or Rag2c/c leukemic cells, respectively. (D) The 24 off-target recombination genes were retrieved by COSMIC Cancer Gene Census (http://cancer.sanger.ac.uk/census/). 0.5 genes and 0.5 cancers gene average sample in Ragf/f, leukemic cells; 8 genes and 4.5 cancer genes average sample in Rag1c/c leukemic cells; 3.3 genes and 0.3 cancer genes average sample in Rag2c/c leukemic cells.
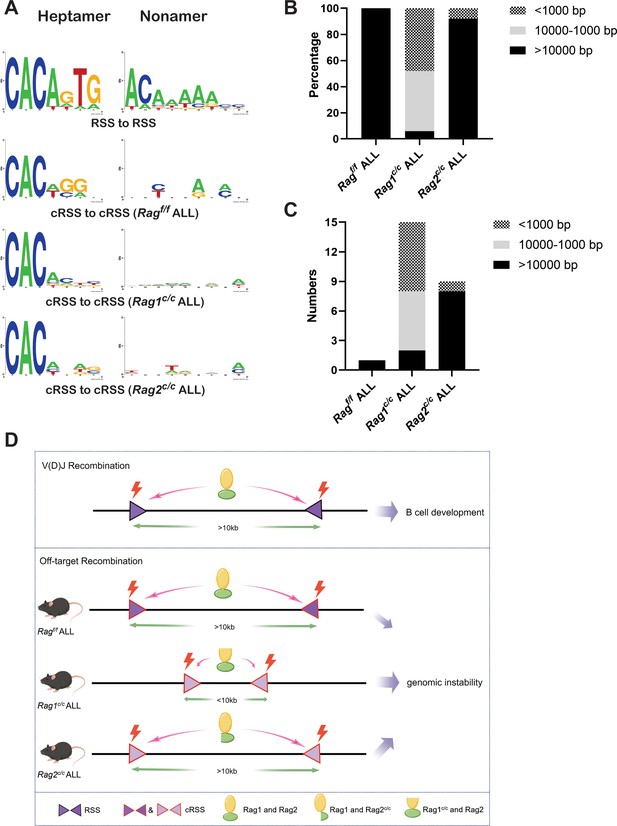
The non-core regions have effects on RAG-binding accuracy and recombinat size in BCR-ABL1+ B lymphocyte.
(A) Sequence logos were used to compare the recombination signal sequence (RSS) and cryptic RSS (cRSS) in Ig loci and non-Ig loci. Top panel: V(D)J recombination at Ig locus; the next three panels: RAG-mediated off-target recombination at non-Ig locus from Ragf/f, Rag1c/c, and Rag2c/c leukemic cells, respectively. The scale of recombinant size was categorized into three ranges: <1000, 1000–10,000, and >10,000 bp. The distribution of different recombinant sizes in Rag f/f, Rag1c/c, and Rag2c/c leukemic cells was presented in (B), while the number of different recombinant sizes in Rag f/f, Rag1c/c, and Rag2c/c leukemic cells is displayed in (C). (D) A schematic depiction of the mechanism of cRAG-accelerated off-target V(D)J recombination was provided. Both RAG1 and RAG2’s non-core region deletion decreases RAG-binding accuracy in Rag1c/c and Rag2c/c, BCR-ABL1+ B-ALL. Additionally, RAG1’s non-core region deletion significantly reduces the size and scale of off-target V(D)J recombination in Rag1c/c BCR-ABL1+ B-ALL.
Additional files
-
Supplementary file 1
Sequencing statistics.
- https://cdn.elifesciences.org/articles/91030/elife-91030-supp1-v1.xlsx
-
Supplementary file 2
Abnormal rearrangements at Ig.
- https://cdn.elifesciences.org/articles/91030/elife-91030-supp2-v1.xlsx
-
Supplementary file 3
RAG-mediated off-target genes.
- https://cdn.elifesciences.org/articles/91030/elife-91030-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91030/elife-91030-mdarchecklist1-v1.pdf