Intermittent fasting promotes type 3 innate lymphoid cells secreting IL-22 contributing to the beigeing of white adipose tissue
Figures
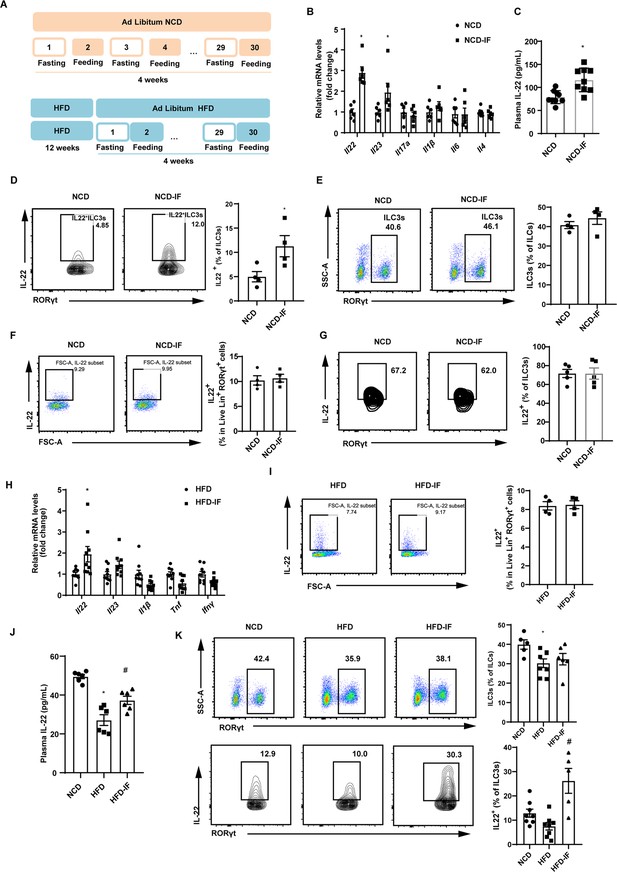
Intermittent fasting enhances interleukin-22 (IL-22) production by intestinal type 3 innate lymphoid cells (ILC3s).
(A) Schematic illustration of the alternate day fasting regimen. NCD, normal chow diet. HFD, high-fat diet. We applied alternate day fasting to mice fed normal chow diet (NCD-IF group) or high-fat diet (HFD-IF group). The control groups were at free access to NCD (NCD group) or HFD (HFD group). n=9 for each group. (B) mRNA expression levels of cytokine genes in the small intestine of NCD and NCD intermittent fasting (NCD-IF) mice. qPCR results were normalized to β-actin. n=6 for each group. (C) Protein levels of IL-22 in plasma of NCD and NCD-IF mice. n=9 for each group. (D) Fl IL-22+ cells in live CD127+ lineage- RORγt+ ILC3s from the small intestine lamina propria (siLP) of NCD and NCD-IF mice. Four independent experiments were performed with similar results. n=4. (E) Flow cytometric analysis of RORγt+ ILC3s in live CD127+ lineage- ILCs in the siLP of NCD and NCD-IF mice. n=4. (F) Flow cytometric analysis of IL-22+ cells in live lineage+ RORγt+ cells in the siLP of mice fed NCD with or without intermittent fasting. n=4. (G) Flow cytometric analysis of IL-22+ cells in CD90.2+ lineage- RORγt+ ILC3s from the stromal vascular fraction (SVF) cells of subcutaneous white adipose tissue (sWAT) in mice fed NCD with or without intermittent fasting. n=5. (H) mRNA expression levels of cytokine genes in the small intestine of mice fed HFD with or without intermittent fasting. qPCR results were normalized to β-actin. n=9. (I) Flow cytometric analysis of IL-22+ cells in live lineage+ RORγt+ cells in the siLP of mice fed HFD with or without intermittent fasting. n=4. (J) Levels of IL-22 in plasma of NCD mice, HFD mice, and HFD-IF mice. n=6. (K) Flow cytometric analysis of RORγt+ ILC3s in live CD127+ lineage- ILCs and flow cytometric analysis of IL-22+ cells in live CD127+ lineage- RORγt+ ILC3s from the siLP of NCD mice, HFD mice, and HFD-IF mice. n=5–8. * vs NCD, # vs HFD, p<0.05. All data represent the mean ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test (A–I) or one-way ANOVA (J and K). NCD, normal chow diet. HFD, high-fat diet. NCD-IF, normal chow diet with intermittent fasting. HFD-IF, high-fat diet with intermittent fasting.
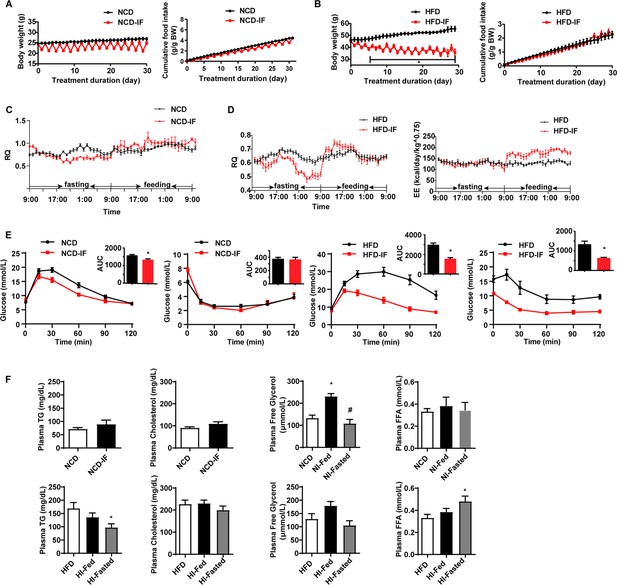
Intermittent fasting improves glucose metabolism and lipid metabolism.
(A) Body weight and cumulative food intake normalized to body weight of mice fed normal chow diet (NCD) with or without intermittent fasting. n=9. (B) Body weight and cumulative food intake normalized to body weight of mice fed high-fat diet (HFD) with or without intermittent fasting. n=9. (C) Respiratory quotient (RQ) of mice fed NCD with or without intermittent fasting. n=5. (D) RQ, energy expenditure (EE) of mice fed HFD with or without intermittent fasting. n=5. (E) Oral glucose tolerance test (OGTT) and area under the curve (AUC). Insulin tolerance test (ITT) and AUC of NCD, NCD-IF, HFD, and HFD-IF mice. n=6 per group. (F) Levels of plasma triglycerides (TG), cholesterol, free glycerol, and free fatty acids (FFA). n=6mice/group. All data represent the mean ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test or one-way ANOVA.
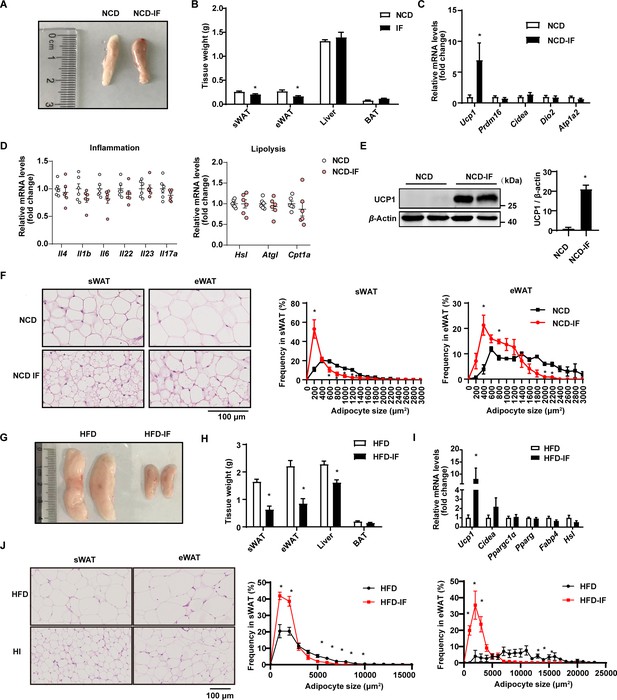
Intermittent fasting promotes white adipose tissue beigeing.
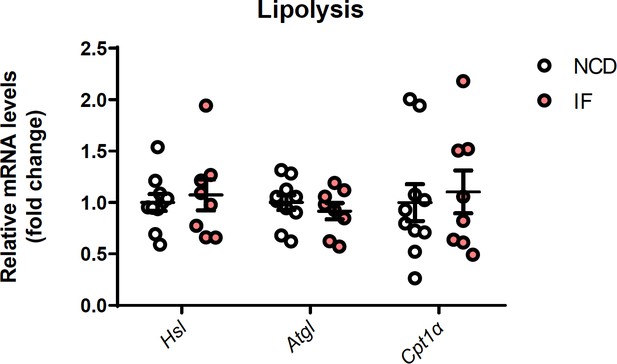
(A) Representative image of subcutaneous white adipose tissue (sWAT) from mice fed normal chow diet (NCD) with or without intermittent fasting. (B) Tissue weight of sWAT, epididymal white adipose tissue (eWAT), liver, BAT, from NCD and NCD intermittent fasting (NCD-IF) mice. n=6. (C) qPCR analysis of thermogenic genes in sWAT from NCD and NCD-IF mice. n=6mice/group. (D) qPCR analysis of inflammation and lipolysis genes in sWAT from NCD and NCD-IF mice. n=6mice/group. (E) UCP1 protein expression in the sWAT of NCD and NCD-IF mice detected by western blotting. β-Actin was used as the loading control. The relative protein signal intensity was quantified using ImageJ software. (F) Hematoxylin-eosin (H&E) staining in sWAT and eWAT. Cell sizes were measured using ImageJ. n=5 per group. Scale bar: 100μm. (G) Representative image for sWAT of HFD and HFD-IF mice. n=6. (H) Tissue weight of sWAT, BAT, liver, eWAT from HFD and HFD-IF mice. n=6. (I) qPCR analysis of thermogenic genes in sWAT from HFD and HFD-IF mice. n=6mice/group. (J) H&E staining in sWAT and eWAT from HFD and HFD-IF mice. Cell sizes were measured using ImageJ. n=5 per group. Scale bar: 100μm. All data represent the mean ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test (B–E, H, I).
-
Figure 1—figure supplement 2—source data 1
Original file for the western blot analysis in Figure 1—figure supplement 2E (anti-UCP1, anti-β-actin).
- https://cdn.elifesciences.org/articles/91060/elife-91060-fig1-figsupp2-data1-v2.zip
-
Figure 1—figure supplement 2—source data 2
PDF containing original scans of the relevant western blot analysis (anti-UCP1, anti-β-actin) with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/91060/elife-91060-fig1-figsupp2-data2-v2.zip
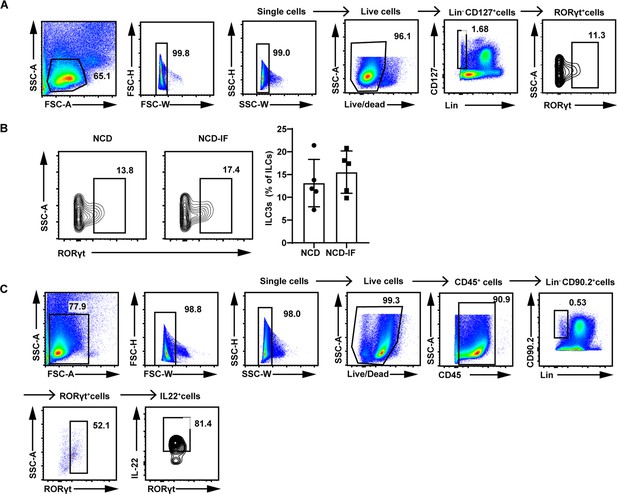
Intermittent fasting demonstrates no effect on the number of type 3 innate lymphoid cells (ILC3s) in subcutaneous white adipose tissue (sWAT).
(A) Gating strategy for flow cytometry analysis of live lineage-CD127+RORγt+ILC3s in the stromal vascular fraction (SVF) of sWAT. (B) Flow cytometric analysis of RORγt+ ILC3s in live CD127+ lineage- ILCs from the sWAT of normal chow diet (NCD) mice, NCD intermittent fasting (NCD-IF) mice. n=5. (C) Gating strategy for flow cytometry analysis of IL-22+ cells in live CD45+lineage-CD90.2+RORγt+ILC3s in the SVF of sWAT. All data represent the mean ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test.
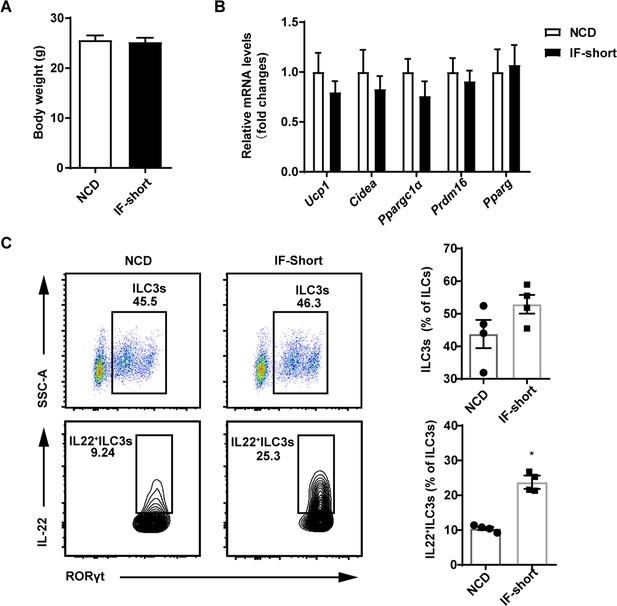
Short-term intermittent fasting induces intestinal type 3 innate lymphoid cells (ILC3s) to secrete interleukin-22 (IL-22).
Eight-week-old SPF mice were exposed to one cycle of intermittent fasting for 2days (IF-short), while the control group (normal chow diet [NCD]) has free access to NCD. n=6. (A) Body weight of NCD and IF-short mice. n=6. (B) qPCR analysis of thermogenic genes in subcutaneous white adipose tissue (sWAT) from NCD and IF-short mice. n=6mice/group. (C) Flow cytometric analysis of RORγt+ ILC3s in live CD127+ lineage- ILCs in the small intestine lamina propria of NCD or IF-short mice. Flow cytometric analysis of IL-22+ cells in live CD127+ lineage- RORγt+ ILC3s from the small intestine lamina propria of NCD or IF-short mice. Four independent experiments were performed with similar results. n=4. All data represent the mean ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test.
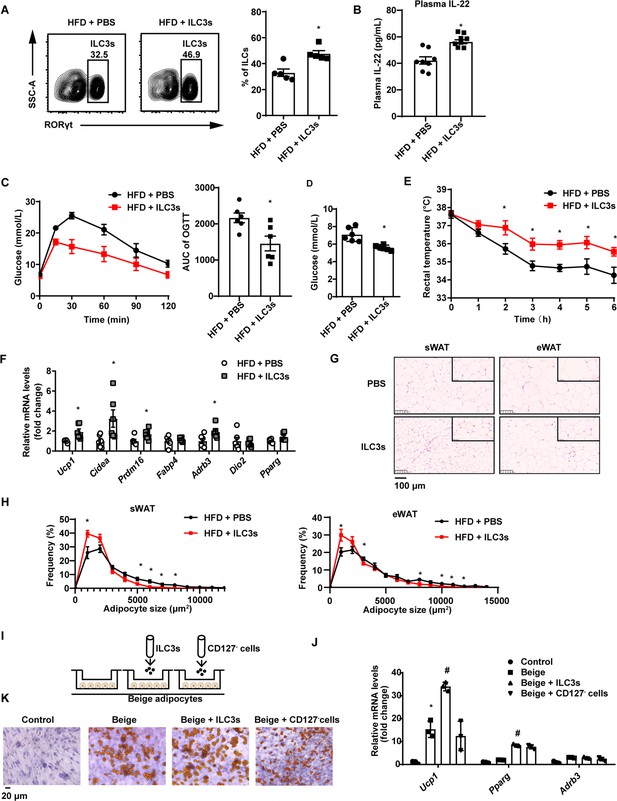
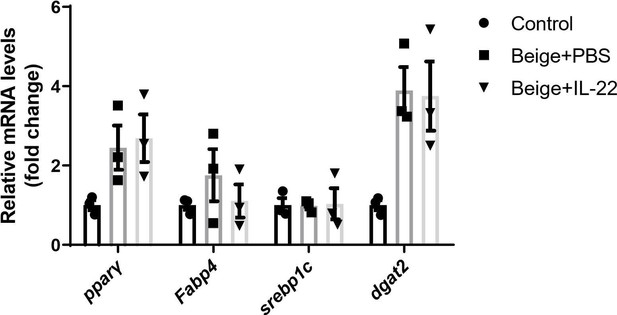
Type 3 innate lymphoid cells (ILC3s) promote beigeing of white adipose tissue.
Six-week-old male C57BL/6J SPF wild-type mice were fed with high-fat diet (HFD) for 16weeks and then injected with ILC3s (HFD + ILC3s group) or phosphate-buffered saline (PBS) (HFD+PBS group) intravenously six times in a month. n=8 for each group. (A) Flow cytometric analysis of RORγt+ ILC3s in live CD127+ lineage- ILCs from the small intestine lamina propria (siLP) of mice transferred with PBS or ILC3s. The proportion of ILC3s in ILCs was shown in the histogram. n=5 for each group. (B) Levels of interleukin-22 (IL-22) in plasma. n=8 for each group. (C) Oral glucose tolerance test (OGTT) and area under the curve (AUC). n=6mice/group. (D) Random blood glucose. n=6mice/group. (E) Rectal temperature of HFD mice transferred with PBS or ILC3s during a 6hr cold challenge (4°C). n=6mice/group. (F) qPCR analysis of thermogenic genes in subcutaneous white adipose tissue (sWAT). n=6mice/group. (G) Representative images of hematoxylin-and-eosin-stained sections of sWAT, epididymal white adipose tissue (eWAT), and BAT from HFD mice transferred with ILC3s or control (n=5). (H) Distribution and average adipocyte size of sWAT and eWAT were shown (n=5). (I) Schematic depicting the co-culture of ILC3s with stromal vascular fraction (SVF)-derived beige adipocytes. (J) qPCR analysis of thermogenic genes in SVF-derived cells. * indicates p<0.05vs control, # denotes p<0.05vs beige. n=3. (K) Oil red O staining of adipocytes after co-culture with ILC3s or CD127- cells. All data represent the means ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test (A–H) or one-way ANOVA (J).
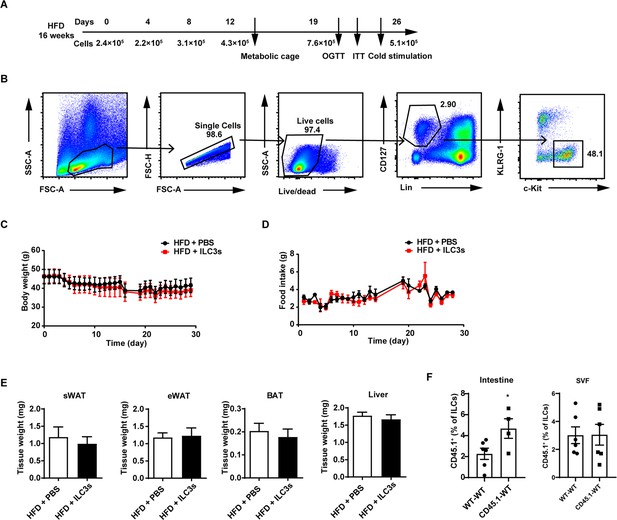
Adoptive transfer of type 3 innate lymphoid cells (ILC3s) has no effect on the body weight in mice fed high-fat diet (HFD).
(A) Timeline for the recipient mice transferred with phosphate-buffered saline (PBS) or ILC3s. Mice were fed with HFD for 16weeks and then injected with ILC3s intravenously six times in a month. (B) Gating strategy for flow sorting of live CD127+ lineage- c-kit+ KLRG1-ILC3s in the small intestine lamina propria (siLP). (C) Body weight of HFD mice transferred with ILC3s or control. n=6 per group. (D) Food intake of HFD mice transferred with ILC3s or control. n=6 per group. (E) Tissue weight of subcutaneous white adipose tissue (sWAT), epididymal white adipose tissue.(eWAT), BAT, and liver from HFD mice transferred with ILC3s or control. n=6 per group. (F) Transfer of ILC3s from CD45.1 mice to wild-type (WT) mice. The percentage of CD45.1+ cells in ILCs in the small intestine and stromal vascular fraction (SVF). n=4–6. All data represent the mean ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test.
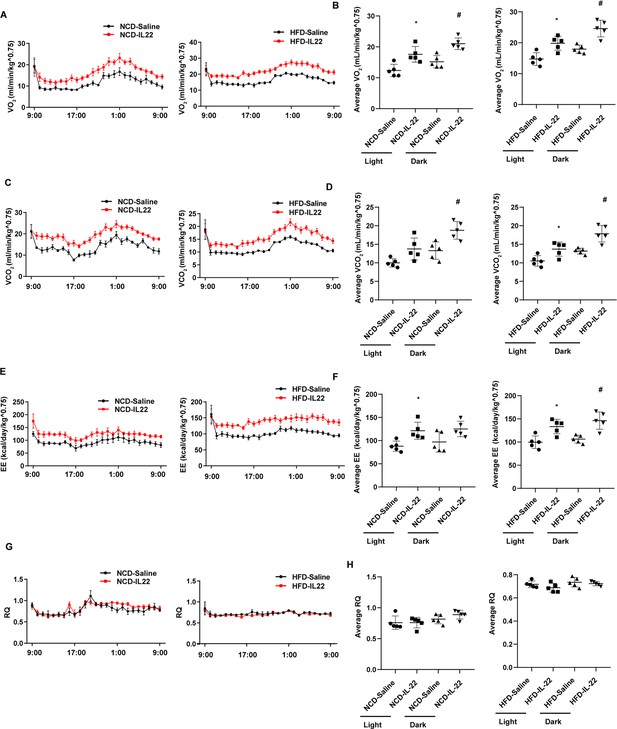
Interleukin-22 (IL-22) promotes energy expenditure.
Six-week-old male C57BL/6J SPF wild-type mice were fed normal chow diet (NCD) or high-fat diet (HFD) for 12 weeks and then divided into four groups (NCD-saline, NCD-IL-22, HFD-saline, HFD-IL-22). Mice were intraperitoneally injected with 4 µg/kg IL-22 every other day for 6 weeks. The control groups were injected with saline. (A) VO2 of mice fed NCD or HFD. (B) Average VO2 at light and dark respectively. (C) VCO2 of mice fed NCD or HFD. (D) Average VCO2 at light and dark respectively. (E) Energy expenditure of mice fed NCD or HFD. (F) Average energy expenditure at light and dark respectively. (G) Respiratory quotient (RQ) of mice fed NCD or HFD. (H) Average RQ at light and dark respectively. * indicates p<0.05 vs NCD-saline or HFD-saline at light. # indicates p<0.05 vs NCD-saline or HFD-saline at dark. n = 5. Statistical significance was determined by one-way ANOVA (B, D, F, H).
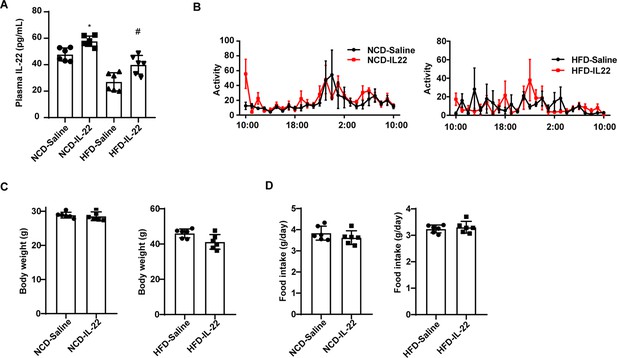
Exogenous interleukin-22 (IL-22) has no effect on the body weight of mice.
(A) Levels of IL-22 in plasma of NCD-saline, NCD-IL-22, HFD-saline, HFD-IL-22 mice. n=6 mice/group. # vs HFD-saline. (B) Activity of mice fed NCD or HFD which were intraperitoneally injected with IL-22 or saline. n=5 for each group. (C) Body weight of NCD and HFD mice intraperitoneally injected with IL-22 or saline. n=6 for each group. (D) Daily food intake of NCD and HFD mice intraperitoneally injected with IL-22 or saline. n=6 for each group. All data represent the mean ± s.e.m. Statistical significance was determined by one-way ANOVA (A) or unpaired two-tailed Student’s t test (C, D). NCD, normal chow diet; HCD, high-fat diet.
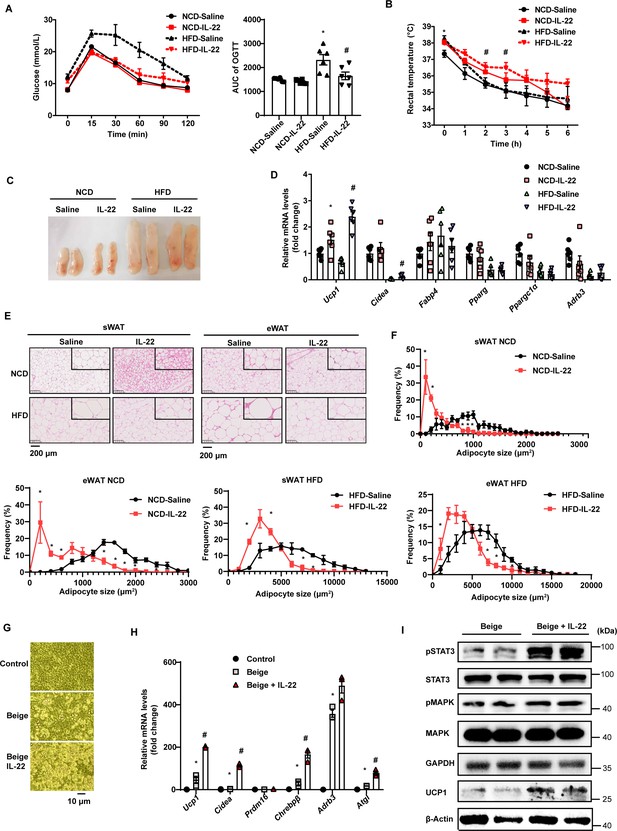
Interleukin-22 (IL-22) promotes beigeing of white adipose tissue.
Six-week-old male C57BL/6J SPF wild-type mice were fed normal chow diet (NCD) or high-fat diet (HFD) for 12weeks and then divided into four groups (NCD-saline, NCD-IL-22, HFD-saline, HFD-IL-22). Mice were intraperitoneally injected with 4µg/kg IL-22 every other day for 6weeks. The saline group was injected with saline. n=6mice/group. (A) Oral glucose tolerance test (OGTT) and area under the curve (AUC). n=6mice/group. * indicates p<0.05vs NCD-saline; # denotes p<0.05vs HFD-saline. (B) Rectal temperature of mice during a 6hr cold challenge (4°C). n=6. * indicates p<0.05vs NCD-saline; # denotes p<0.05vs HFD-saline. (C) Representative image of sWAT of the four groups, NCD-saline, NCD-IL-22, HFD-saline, HFD-IL-22. (D) qPCR analysis of thermogenic genes of sWAT. n=6mice/group. * indicates p<0.05vs NCD-saline; # denotes p<0.05vs HFD-saline. (E) Representative images of hematoxylin-and-eosin-stained sections of sWAT and eWAT (n=5 for each group). (F) The distribution and average adipocyte size of sWAT and eWAT were determined by ImageJ. (G) Phase-contrast microscopic images of stromal vascular fraction (SVF) cells and adipocytes. (H) qPCR analysis of thermogenic genes in SVF cells and beige adipocytes. n=3. # denotes p<0.05vs beige. (I) pSTAT3, STAT3, pMAPK, MAPK, GAPDH, UCP1, β-actin protein expression in the beige adipocytes or beige adipocytes treated with IL-22 detected by western blotting. GAPDH and β-actin was used as the loading control. All data represent the mean ± s.e.m. Statistical significance was determined by one-way ANOVA (A–D, H) or two-tailed Student’s t test (F). sWAT, subcutaneous white adipose tissue; eWAT, epididymal white adipose tissue.
-
Figure 4—source data 1
Original file for the western blot analysis in Figure 4I (anti-pSTAT3, anti-STAT3, anti-pMAPK, anti-MAPK, anti-GAPDH, anti-UCP1, anti-β-actin).
- https://cdn.elifesciences.org/articles/91060/elife-91060-fig4-data1-v2.zip
-
Figure 4—source data 2
PDF containing original scans of the relevant western blot analysis (anti-pSTAT3, anti-STAT3, anti-pMAPK, anti-MAPK, anti-GAPDH, anti-UCP1, anti-β-actin) with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/91060/elife-91060-fig4-data2-v2.zip
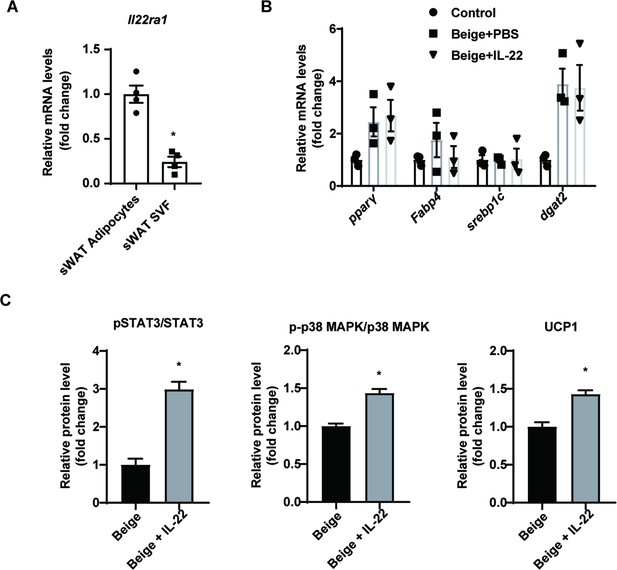
Interleukin-22 (IL-22) can directly act on adipocytes.
(A) qPCR analysis of Il22ra1 genes in adipocytes or stromal vascular fraction (SVF) cells isolated from subcutaneous white adipose tissue (sWAT). n=4. * denotes p<0.05vs sWAT adipocytes. (B) qPCR analysis of adipogenic marker genes of SVF cells and adipocytes. n=3. (C) Relative protein signal intensity quantified using ImageJ software. All data represent the mean ± s.e.m. Statistical significance was determined by one-way ANOVA (B) or unpaired two-tailed Student’s t test (A, C).
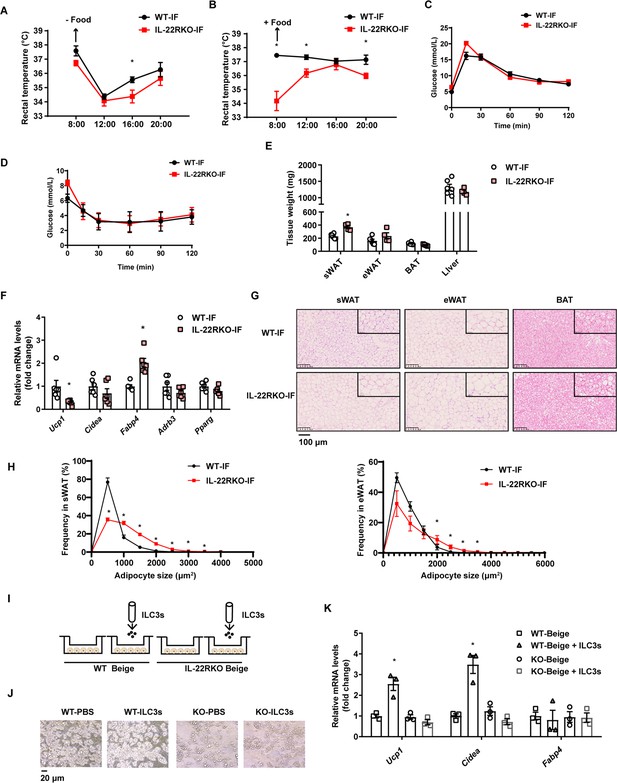
Interleukin-22R knockout (IL-22RKO) blocks white adipose tissue beigeing induced by intermittent fasting.
Eight-week-old IL-22RKO and wild-type (WT) mice were subjected to alternate day fasting for 30days. n=6mice per group. (A) Rectal temperature of mice at room temperature during the fasting day. n=6 per group. (B) Rectal temperature of mice at room temperature during the fed day. n=6 per group. (C) Oral glucose tolerance test (OGTT) of WT-IF mice and IL-22RKO-IF mice. n=6 per group. (D) Insulin tolerance test (ITT) of WT-IF mice and IL-22RKO-IF mice. n=6 per group. (E) Tissue weight of subcutaneous white adipose tissue (sWAT), epididymal white adipose tissue (eWAT), BAT, and liver from WT-IF and IL-22RKO-IF mice. n=6mice/group. (F) qPCR analysis of thermogenic genes in sWAT from WT-IF and IL-22RKO-IF mice. n=6mice/group. (G) Representative images of hematoxylin-and-eosin-stained sections of sWAT, eWAT, and BAT (n=5 for each group). (H) The distribution and average adipocyte size of sWAT and eWAT were determined by ImageJ. (I) Schematic depicting the co-culture of type 3 innate lymphoid cells (ILC3s) with stromal vascular fraction (SVF)-induced beige adipocytes from WT or IL-22RKO mice. n=3. Experiments were repeated three times. (J) Phase-contrast microscopy images of beige adipocytes differentiated from SVF cells co-cultured with or without ILC3s. Shown are representatives from one experiment. (K) qPCR analysis of thermogenic genes in SVF-derived cells co-cultured with or without ILC3s. n=3. All data represent the means ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test (A–H) or one-way ANOVA (K).
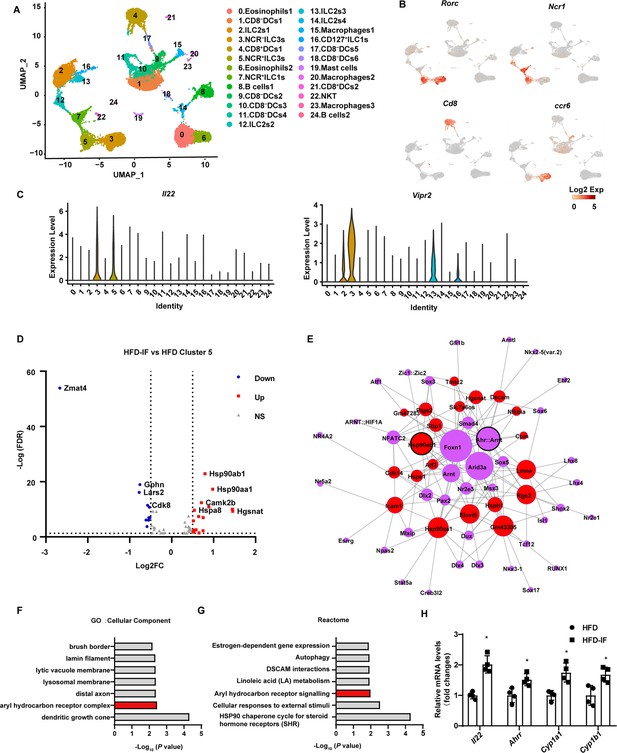
Profiling of intestinal immune cells derived from mice fed normal chow diet (NCD), high-fat diet (HFD), and high-fat diet with intermittent fasting (HFD-IF).
Live CD45+ lineage-cells sorted from mice fed normal chow diet (NCD group), high-fat diet (HFD group), high-fat diet with alternate day fasting (HFD-IF group) were analyzed using single-cell sequencing. (A) Cell subsets in the small intestine lamina propria CD45+ lineage- immune cell atlas. Two-dimensional (2D) representation of cell profiles (dots) from the small intestine lamina propria, colored and numbered by cluster membership. (B and C) UMAP feature plots (B) and violin plots (C) showing RNA expression of cluster markers for the indicated cell populations. UMAP feature plots are based on the UMAP shown in (A). (D) The volcano plot of differentially expressed genes in cluster 5 (NCR+ type 3 innate lymphoid cells [ILC3s]). The red dots represent upregulated genes in HFD-IF group compared with HFD group, while the blue dots represent downregulated genes in HFD-IF group compared with HFD group. Hsp90ab1 is one of the notably upregulated genes. (E) Transcription factor prediction using the Jaspar database and TFBS tools. Red dots represent differentially expressed genes, purple dots represent transcription factors, and larger nodes represent more nodes connected to them. (F) Gene ontology (GO) of the top 20 differentially expressed genes in cluster 5. The cellular component up GO terms in HFD-IF group compared with HFD group. (G) The reactome up terms of cluster 5 top 20 differential genes in HFD-IF group compared with HFD group. (H) qPCR analysis of AHR target genes in ILC3s sorted from HFD and HFD-IF mice. n=4. *, p<0.05. All data represent the means ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test.
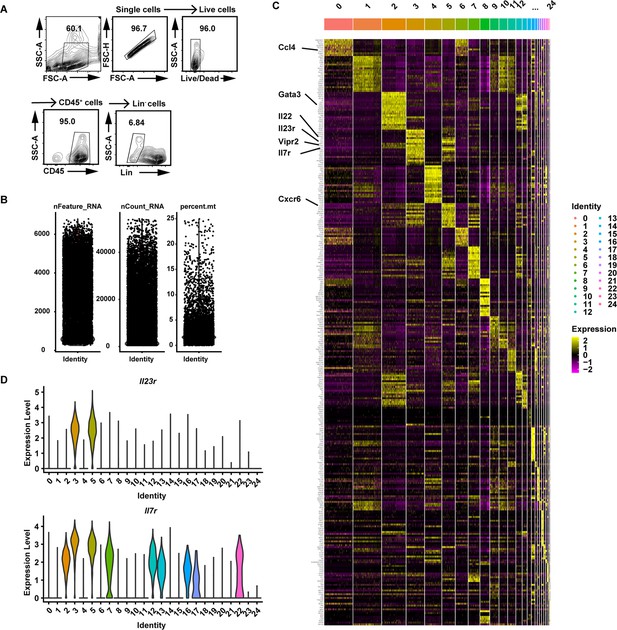
Profiling of intestinal immune cells from mice fed normal chow diet (NCD), high-fat diet (HFD), or high-fat diet with intermittent fasting (HFD-IF).
(A) Gating strategy for flow cytometry sorting of live CD45+ lineage- immune cells in the small intestine lamina propria. (B) scRNA-seq data quality control of sorted immune cells from mice fed NCD, HFD, or HFD-IF. (C) Unbiased heatmap of the gene levels of the top 20 unique cluster marker genes for each cell cluster. Cluster identities are shown above the heatmap. (D) Violin plots showing RNA expression of cluster markers for the indicated cell populations. UMAP feature plots are based on the UMAP shown in Figure 6A.
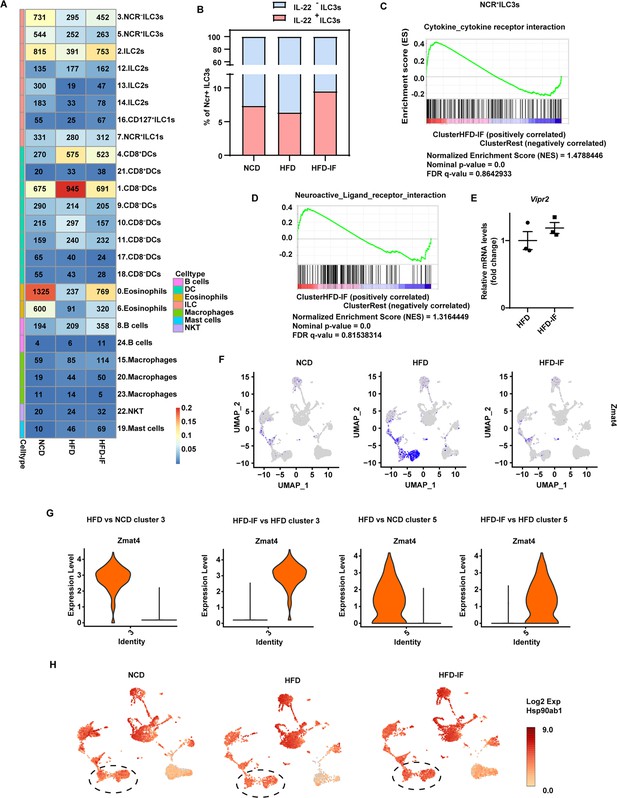
Effects of intermittent fasting on the gene expression of type 3 innate lymphoid cells (ILC3s).
(A) Number and proportion of cells detected in each group in the study. Shown are fraction (color bar, out of all cells from that group) and number of cells from each of the 25cell clusters (rows) in each individual group (columns). (B) Percentage of IL-22+ cells defined by the mRNA levels using Loupe Browser 5.0 of NCR+ ILC3s in normal chow diet (NCD), high-fat diet (HFD), and high-fat diet with intermittent fasting (HFD-IF) mice. (C) GSEA demonstrating enrichment of cytokine-cytokine receptor interaction pathway genes upregulated by intermittent fasting (HFD-IF mice) vs. control (HFD mice) in NCR+ ILC3s. (D) GSEA demonstrating enrichment of neuroactive ligand-receptor interaction pathway genes upregulated by intermittent fasting (HFD-IF mice) vs. control (HFD mice) in NCR+ ILC3s. (E) qPCR analysis of Vipr2 in ILC3s sorted from small intestine lamina propria (siLP) of HFD or HFD-IF mice. (F) UMAP feature plots showing RNA expression of Zmat4 based on the UMAP shown in Figure 6B. (G) Violin plots showing RNA expression of Zmat4 in NCR-ILC3s (cluster 3) and NCR+ILC3s (cluster 5). (H) UMAP feature plots showing RNA expression of Hsp90ab1 based on the UMAP shown in Figure 6B.
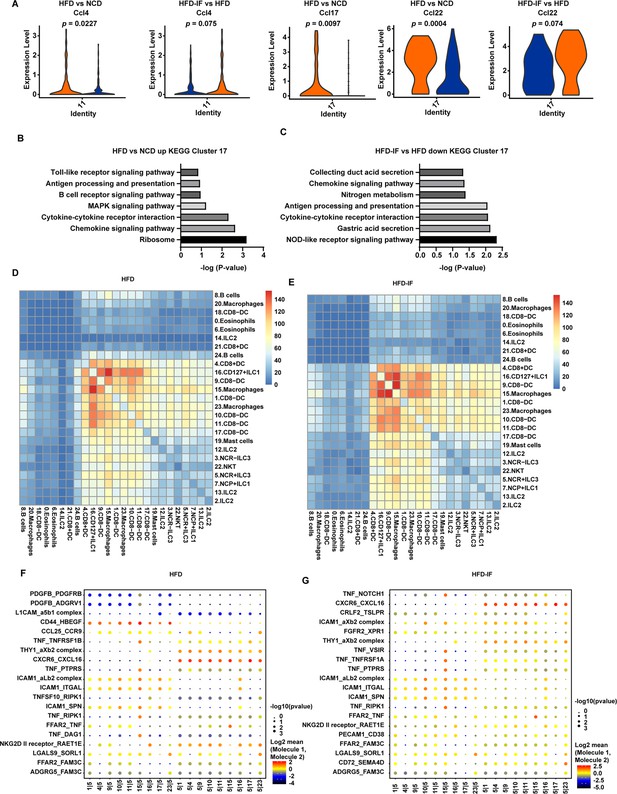
Interaction between myeloid cells and type 3 innate lymphoid cells (ILC3s).
(A) Violin plots showing the RNA expression of chemokines in CD8- dendritic cells (DCs) (cluster 11 and cluster 17). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment in upregulated genes of CD8-DCs of high-fat diet (HFD) group compared with normal chow diet (NCD) group (cluster 17). (C) KEGG enrichment of downregulated genes of CD8-DCs of high-fat diet with intermittent fasting (HFD-IF) group compared with HFD group (cluster 17). (D) Heatmap showing the number of significant interactions identified between cell types in sorted small intestine lamina propria (siLP) immune cells of HFD mice as determined by CellPhoneDB. The color represents the number of interactions between cell types, a higher number of interactions (red), and a lower number of interactions (blue). (E) Heatmap showing the number of significant interactions identified between cell types in sorted siLP immune cells of HFD-IF mice as determined by CellPhoneDB. The color represents the number of interactions between cell types: a higher number of interactions (red) and a lower number of interactions (blue). (F) Interaction pattern of the top 20 protein pairs and the top 20cell types in sorted siLP immune cells of HFD group mice. The x-axis is the cell-type interaction, and the y-axis is the protein interaction. The larger the point is, the smaller the p value. The color represents the average expression, and red to black indicates the level from high to low. (G) Interaction pattern of the top 20 protein pairs and the top 20cell types in sorted siLP immune cells of HFD-IF mice. The x-axis is the cell-type interaction, and the y-axis is the protein interaction. The larger the point is, the smaller the p value. The color represents the average expression, and red to black indicates the level from high to low.
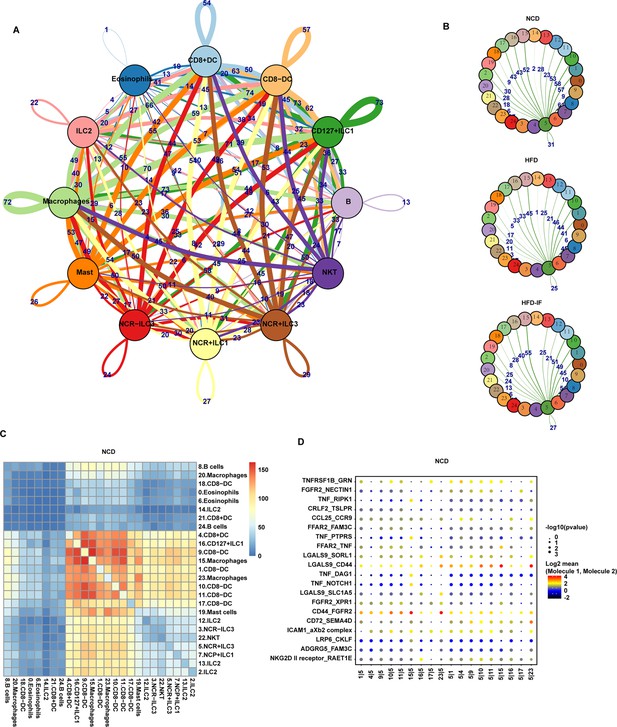
Cell-cell interactions in the intestine.
(A) Connectome web analysis of intestine immune cell interacting cell types based on the expression of the ligand in mice fed normal chow diet (NCD). The vertex (colored cell node) represents the cell cluster. The thickness of the connecting lines is proportional to the number of interactions between the nodes. (B) Connectome web analysis of NCR+ type 3 innate lymphoid cells (ILC3s) with the 25cell clusters based on expression of the ligand. The vertex (colored cell node) represents the cell cluster, while the thickness of the connecting lines is proportional to the number of interactions between two nodes. (C) Heatmap showing the number of significant interactions identified between cell types in sorted small intestine lamina propria (siLP) immune cells of NCD mice as determined by CellPhoneDB. The color represents the number of interactions between cell types: a higher number of interactions (red) and a lower number of interactions (blue). (D) Interaction pattern of the top 20 protein pairs and the top 20cell types. The x-axis is the cell type interaction, and the y-axis is the protein interaction. The larger the point is, the smaller the p value. The color represents the average expression. Red to black indicates the level from high to low.
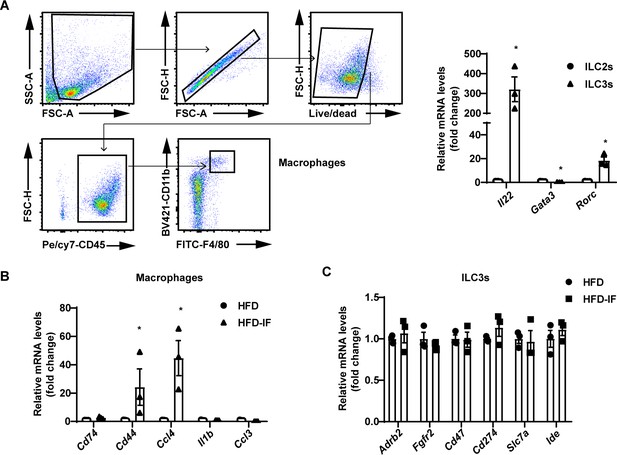
Increased expression of CD44 and CCl4 in macrophages.
Flow cytometry-sorted macrophages and type 3 innate lymphoid cells (ILC3s) from high-fat diet (HFD) and high-fat diet with intermittent fasting (HFD-IF) mice were used to detect the mRNA levels of proteins involved in the interaction of macrophages and ILC3s. (A) Gating strategy for flow sorting of macrophages in the small intestine lamina propria (siLP). qPCR analysis of Il22, Gata3, and Rorc in ILC2s and ILC3s. n=3/group. (B) mRNA levels of proteins involved in the interaction and Il23 in macrophages. n=3/group. (C) mRNA levels of proteins involved in the interaction in ILC3s. n=3/group. All data represent the mean ± s.e.m. Statistical significance was determined by unpaired two-tailed Student’s t test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | PerCP/Cy5.5 anti-mouse CD45(30-F11) (Mouse Monoclonal) | BioLegend | Cat# 103131 | FACS (1:400) |
| Antibody | FITC anti-mouse CD3ε(RA3-6B2) (Rabbit Monoclonal) | BioLegend | Cat# 103205 | FACS (1:400) |
| Antibody | FITC anti-mouse/human CD45R/B220(RA3-6B2) (Rabbit Monoclonal) | BioLegend | Cat# 103205 | FACS (1:400) |
| Antibody | FITC anti-mouse Ly-6G/Ly-6C(Gr-1)(RB6-8C5) (Rabbit Monoclonal) | BioLegend | Cat# 108405 | FACS (1:400) |
| Antibody | FITC anti-mouse CD19(6D5) (Rabbit Monoclonal) | BioLegend | Cat# 115506 | FACS (1:400) |
| Antibody | FITC anti-mouse CD5(53–7.3) (Rabbit Monoclonal) | BioLegend | Cat# 100605 | FACS (1:400) |
| Antibody | BV421 anti-mouse CD127(IL-7Rα) (A7R34) (Rabbit Monoclonal) | BioLegend | Cat# 135023 | FACS (1:400) |
| Antibody | PE/Cyanine7 anti-mouse CD90.2(30-H12) (Rabbit Monoclonal) | BioLegend | Cat# 105325 | FACS (1:400) |
| Antibody | PE/Cyanine7 anti-mouse CD45.1 (Mouse Monoclonal) | BioLegend | Cat# 110729 | FACS (1:400) |
| Antibody | PE anti-mouse RORγt(Q31-378) (Mouse Monoclonal) | BD Pharmingen | Cat# 562607 | FACS (1:400) |
| Antibody | Alexa Fluor 647 anti-mouse IL-22(Poly5164) (Mouse Polyclonal) | BioLegend | Cat# 516406 | FACS (1:400) |
| Antibody | BV605 anti-mouse/human KLRG1(MAFA)(2F1/KLRG1) (Syrian Hamster Monoclonal) | BioLegend | Cat# 138419 | FACS (1:400) |
| Antibody | PE anti-mouse CD117(c-kit) (2B8) (Rabbit Monoclonal) | BioLegend | Cat# 105807 | FACS (1:400) |
| Antibody | Rb polyclonal antibody to UCP1 (Rabbit Polyclonal) | abcam | Cat# ab10983 | WB (1:1000) |
| Antibody | Phospho-Stat3 (Tyr705) Rabbit mAb (Rabbit Monoclonal) | Cell Signaling Technology | Cat# 9145 | WB (1:1000) |
| Antibody | Stat3 (124H6) Mouse mAb (Mouse Monoclonal) | Cell Signaling Technology | Cat# 9139 | WB (1:1000) |
| Antibody | Phospho-p38 MAPK (Thr180/Tyr182) (D3F9) XP Rabbit mAb (Rabbit Monoclonal) | Cell Signaling Technology | Cat# 4511 | WB (1:1000) |
| Antibody | p38 MAPK (D13E1) XP Rabbit mAb (Rabbit Monoclonal) | Cell Signaling Technology | Cat# 8690 | WB (1:1000) |
| Antibody | Rb polyclonal antibody to UCP1 (Rabbit Polyclonal) | abcam | Cat# ab10983 | WB (1:1000) |
| Commercial assay or kit | eBioscience Fixation/Perm Diluent | Invitrogen | Cat# 00-8333-56 | |
| Commercial assay or kit | eBioscience Fixable Viability Dye eFluorTM 606 | Invitrogen | Cat# 65-0866-14 | FACS (1:400) |
Additional files
-
Supplementary file 1
Sequences of primers used in quantitative PCR.
- https://cdn.elifesciences.org/articles/91060/elife-91060-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91060/elife-91060-mdarchecklist1-v2.docx