Near-perfect precise on-target editing of human hematopoietic stem and progenitor cells
Figures
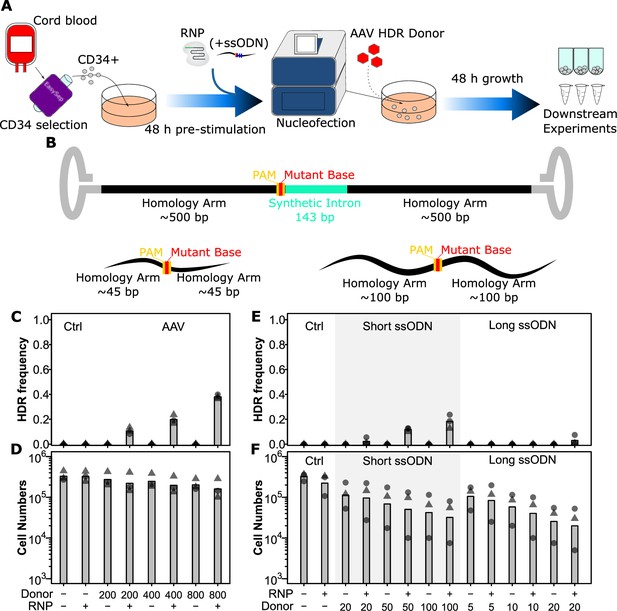
AAV and short ssODN both allow precise editing in human hematopoietic stem and progenitor cells (HSPCs).
(A) Experimental design. (B) Homology-directed repair (HDR) donor configurations. The SRSF2 P95H AAV donor is shown above and short and long ssODN donors are shown below with features indicated. Annotated sequences are shown in Supplementary file 2. (C) HDR integration efficiency by AAV dose. Cells were edited with 30.5 pmol ribonuclear protein (RNP) (or not as indicated) with indicated multiplicities of infection (MOI) of AAV donor. Bars show mean values and points show measurements for individual cords. Male cords are shown as triangles and females as circles. (D) Viable cell number by AAV dose. Hemocytometer counts at the time of harvest are shown for each sample from (C). (E) HDR integration efficiency for short and long ssODN donors. Donor DNA amounts are shown in pmol. (F) Viable cell number by ssODN dose. False-discovery rate (FDR) corrected paired t-test significance values are shown in Supplementary file 1. See also Figure 1—figure supplements 1 and 2.
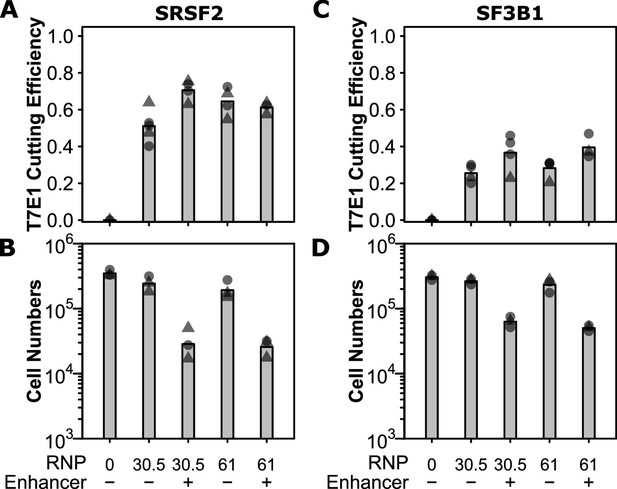
Ribonuclear protein (RNP) efficiency determination.
Cells were edited with indicated amounts of Cas9 RNP and 1 x IDT electroporation enhancer where indicated. Cutting efficiencies as measured by T7E1 are shown in (A) for SRSF2 and (C) for SF3B1, and number of viable cells in (B) for SRSF2 and (D) for SF3B1. False-discovery rate (FDR) corrected unpaired t-test significance values are shown in Supplementary file 1.
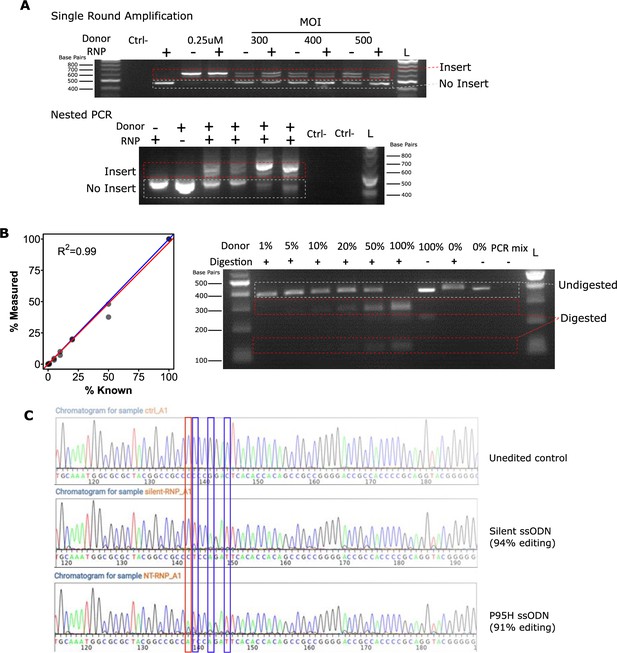
Assays for the detection of homology-directed repair (HDR) integration.
(A) Nested PCR is required to avoid non-specific amplification. The upper gel shows that even in the absence of ribonuclear protein (RNP), single primer pairs amplify AAV from the donor. The lower gel demonstrates that with nested PCR this is no longer the case. (B) Enzymatic digestion can detect editing down to a minority of ~1%. Known proportions of mutant and wild-type PCR products were mixed, digested with BspEI, and the gels quantified. An example gel is shown on the right, and the quantifications from two independent replicates shown on the left. A perfect measure is shown as a blue line and the linear fit in red. (C) Sanger sequencing-based detection of editing. An example chromatogram from an unedited control, a silent edited sample, and a P95H with spacer-breaking silent mutations edited sample are shown. Locations of mutant bases are indicated with a red box for the location of the P95H mutation, and blue boxes for each of the silent mutations. Editing efficiencies calculated by ICE are indicated for each sequence.
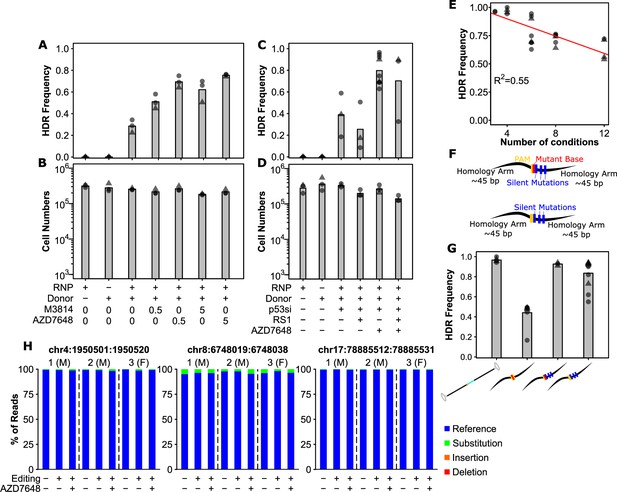
Small molecule-mediated inhibition of DNA-PK and optimal donor design substantially improve precise editing efficiency.
(A) AZD7648 and M3814 improve homology-directed repair (HDR) efficiency in primary human hematopoietic stem and progenitor cell (HSPC). Cells were edited with 30.5 pmol ribonuclear protein (RNP) (or not as indicated) with 400 multiplicity of infection (MOI) of AAV donor and small molecules added as indicated (in µM). Bars show mean values and points show measurements for individual cords. Male cords are shown as triangles and females as circles. (B) Viable cell numbers with AZD7648 and M3814 addition. Hemocytometer counts at the time of harvest are shown for each sample from (A). (C) HDR efficiency with combinations of AZD7648, p53 siRNA, and RS-1. Cells were edited with 30.5 pmol RNP (or not as indicated) with 400 MOI of AAV donor in the presence of the indicated additives. AZD7648 was used at 5 µM, p53 siRNA at 20 fmol, and RS-1 at 15 µM. (D) Viable cell numbers with additive combinations. Hemocytometer counts at the time of harvest are shown for each sample from (C). (E) Technical factors associated with high sample number is associated with decreased HDR efficiency. HDR efficiency is shown for all 30.5 pmol RNP, 400 MOI AAV, 20 fmol p53 siRNA, and 5 µM AZD7648 samples by the number of conditions processed in a given experiment. A linear fit is indicated as a red line. The R2 is indicated, and overall p-value was <<0.001. (F) Alternative designs for ssODN donors with key features indicated. Annotated sequences are shown in Supplementary Information. (G) Silent mutations allow ssODN donors to achieve similar efficiencies to AAV. All edits were performed with 0.5 µM AZD7648, 20 fmol p53 siRNA, 50 pmol ssODN, or 400 MOI AAV as indicated. Donor types are shown as their logos from (1B, 2F). (H) No observable off-target mutations at predicted target sites even with the addition of AZD7648. The overall percent of reads containing exclusively reference allele, or any substitutions, deletions, or insertions that overlap with the predicted off-target cut sites is shown for three individual cords across the top 3 cut sites. Cells from each individual cord were split into an unedited control, and cells edited with the silent mutation containing ssODN for the SRSF2 locus under either standard conditions (i.e. no p53siRNA or AZD7648) or with our optimal editing protocol (i.e. with p53siRNA and 0.5 µM AZD7648). False-discovery rate (FDR) corrected paired t-test significance values are shown in Supplementary file 1. See also Figure 2—figure supplements 1–3.

The addition of AZD7648 also improves editing efficiency at the SF3B1 locus.
(A) Homology-directed repair (HDR) efficiency for SF3B1 K700E with combinations of AZD7648, p53 siRNA and RS-1. Cells were edited with 30.5 pmol ribonuclear protein (RNP) (or not as indicated) with 400 multiplicity of infection (MOI) of AAV donor in the presence of the indicated additives. AZD7648 was used at 5 µM, p53 siRNA at 20 fmol, and RS-1 at 15 µM. (B) Viable cell numbers with additive combinations. Hemocytometer counts at the time of harvest are shown for each sample from (A). False-discovery rate (FDR) corrected unpaired t-test significance values are shown in Supplementary file 1.
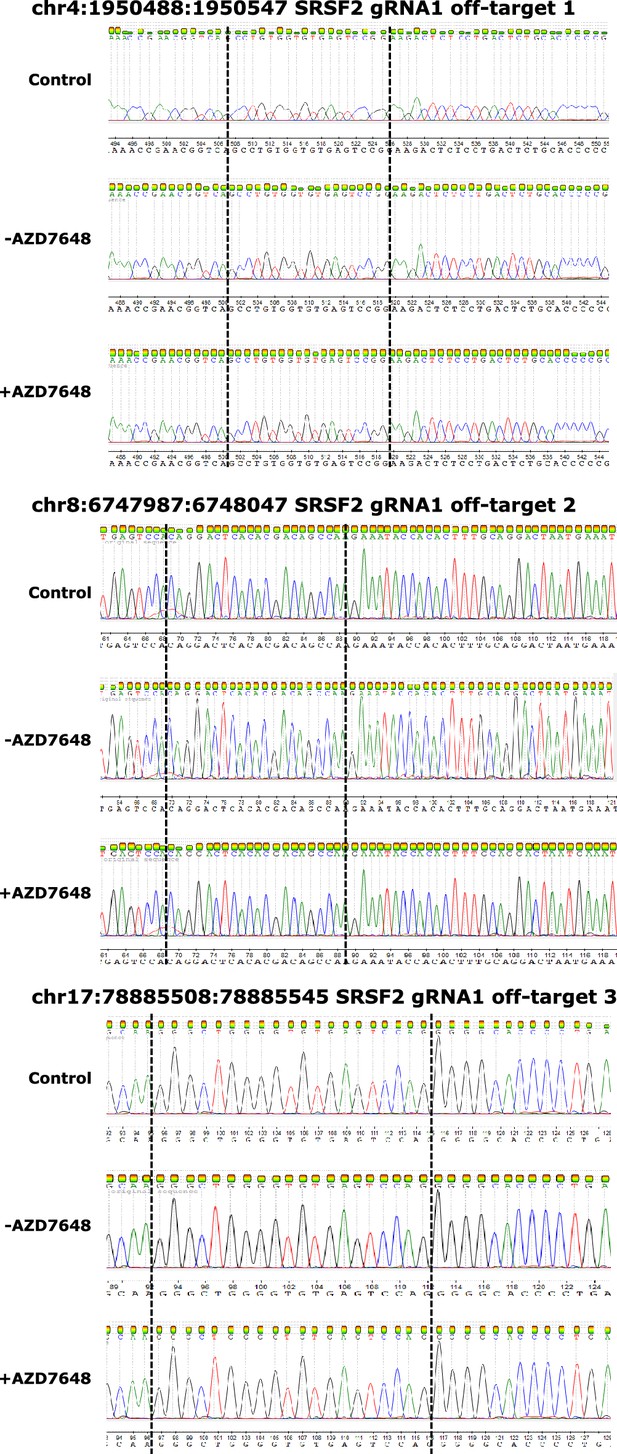
Example Sanger sequencing traces for the top three predicted off-target sites of the SRSF2 gRNA.
Chromosome number and location displayed are indicated for each site. Dotted lines indicate the expected recognition site based on predictions from Benchling. Off-target site 2 is displayed from the reverse strand. All traces were analyzed using ICE and had an estimated cutting of 0% with R2 values between 0.99 and 1. One example chromatogram for each site is shown, however, three independent donors were analyzed for each. Trace views were exported using Chromas.
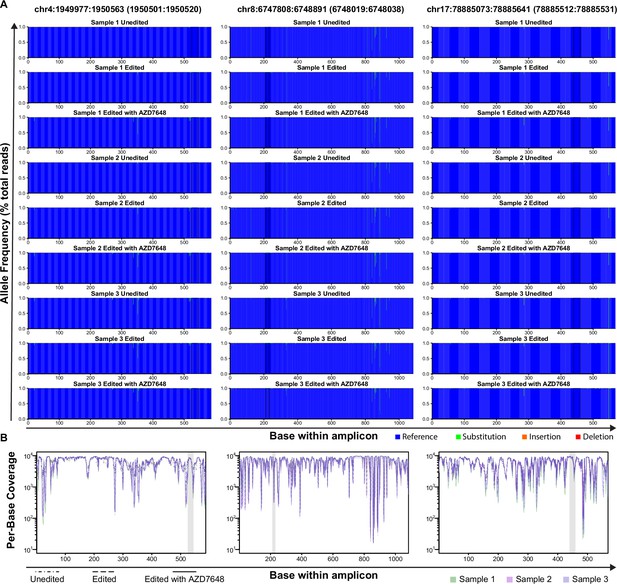
Full-length nanopore sequencing traces for the top three predicted off-target sites of the SRSF2 gRNA.
(A) Frequency of each base type across off-target amplicons. Chromosome number and location are displayed above each column of samples with the overall amplicon first and the specific predicted target location in brackets. The frequency of reads at a given base that were either reference allele, a substitution (from reference), an insertion, or a deletion are shown for each base across the amplicon. Only reads with a Phred score of at least 16 at a given base were included. Results are shown for unmanipulated control, standard editing (ie, no AZD7648), and edited with our optimal protocol (including AZD7648) are shown for each of three individual cord donors (Samples 1–3). Of these, two were male (Samples 1 & 2) and one female (Sample 3). (B) Read depth Per Individual across each amplicon. The total read depth of accepted bases (ie. those with a Phred score of at least 16) is shown for each sample and region. Target regions are highlighted in gray.
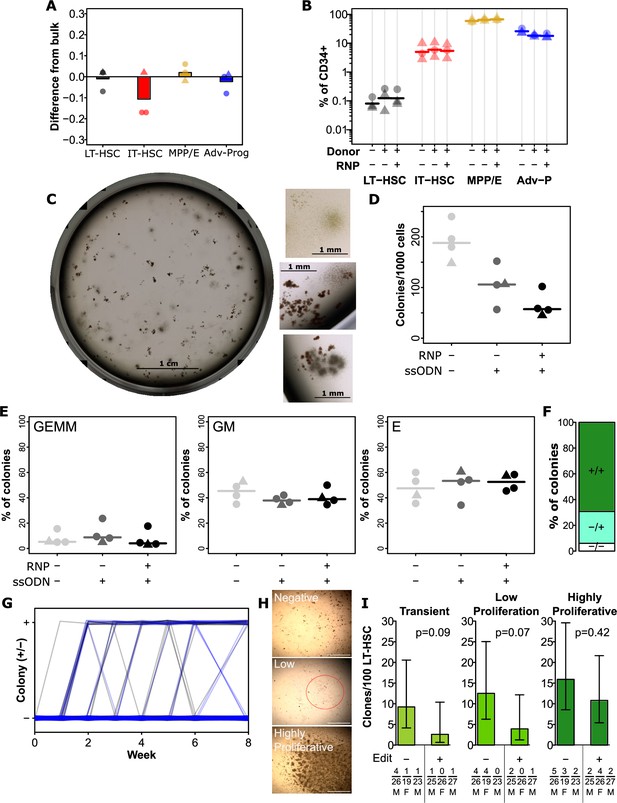
Editing has a minimal impact on hematopoietic stem and progenitor cell (HSPC) function and hierarchy.
(A) Integration efficiency is equivalent across phenotypically defined progenitor compartments. All edits were performed with 0.5 µM AZD7648, 20 fmol p53 siRNA, and 50 pmol of silent mutation ssODN. Values show the difference in precise edit efficiency for each phenotypic subset compared to bulk assessment within that cord. Bars show mean values and points show measurements for individual cords. Male cords are shown as triangles and females as circles. All populations show no significant difference from bulk. (B) Progenitor phenotypes are minimally altered across the hierarchy. The % of CD34 + for each sub-population is shown. Mean values are indicated as lines. A slight but significant decrease was present for late progenitors (CD34 +CD45RA+) associated with donor addition (but not different with editing). (C) An example image of a well of colonies, and example colonies. (D) Total colonies are decreased by the addition of donors, and further by editing. Total CFC per 1000 CD34 + cells is shown for each cord. Lines indicate mean values. (E) No changes were observed in the frequency of colonies of each type. As before points are individual cords and lines show mean values. (F) Colonies showed a preponderance of homozygous editing. Mean homozygous, heterozygous edited, and unedited cells are shown from 36 analyzed colonies across three independent cords. False-discovery rate (FDR) corrected paired t-test significance values are shown in Supplementary file 1. (G) No change in the dynamics of colony emergence from single-LT-HSCs in long-term culture initiating cell (LTC-IC). The presence or absence of an obvious colony in each well (initially sorted with a single long-term HSC (LT-HSC)) was scored weekly over the first 6 weeks of the LTC-IC assay, and again at week 8. Clonal outputs are shown as lines with unedited in black and edited in blue. (H) Example colonies at 8 weeks. At 8 weeks, clones were scored as negative (no colony at any point), transient (previous colony without a colony at endpoint), low proliferation (>50 cells, but below confluence), and highly proliferative (confluent). Example images of negative, low proliferation, and highly proliferative clones are shown. Scalebars (white) show 1 mm. The low proliferation colony is circled in red. (I) Highly proliferative clones are not lost from the LT-HSC population in the editing process. The frequency of clones of the indicated types is shown per 100 phenotypic LT-HSC either without editing or following optimal editing. Error bars represent 95% confidence intervals. Frequencies, p-values, and error bars were calculated using Extreme Limiting Dilution Analysis, based on colony numbers measured from three independent experiments (each with a different cord donor). Numbers for each clone per donor, the total number of clones analyzed for that donor, and donor sex are indicated below the relevant bar. See also Figure 3—figure supplement 1.
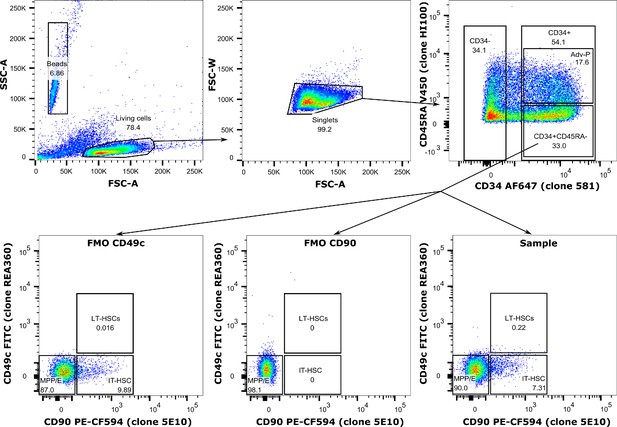
Example gating hierarchy.
FMO stands for full-minus-one, where the indicated antibody was left out of the panel.
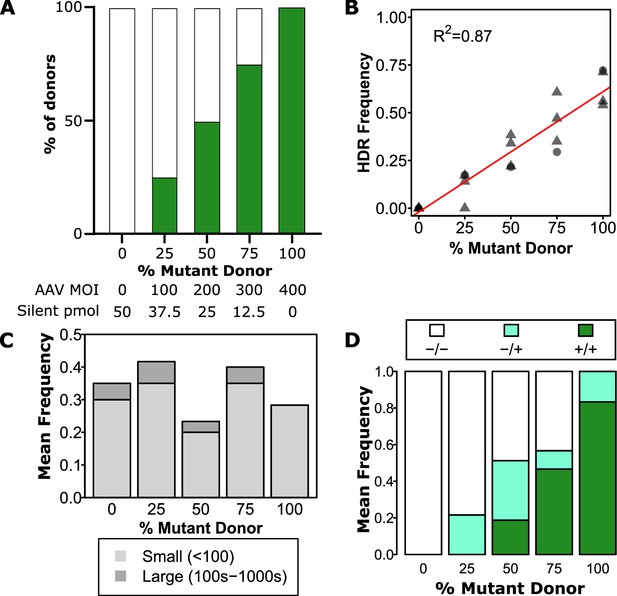
Zygosity can be tuned using a mixture of mutant and silent donors.
(A) Experimental design. Green bars represent the proportion of mutant donor. Specific amounts of mutant and silent donor are shown underneath for each condition. All edits were performed with 0.5 µM AZD7648, 20 fmol p53 siRNA, and indicated amounts of each donor. (B) Overall mutant integration efficiency varies linearly with the proportion of mutant donor. Individual cords are shown as points. Male cords are shown as triangles and females as circles. A linear fit is indicated as a red line. The R2 is indicated, and overall p-value was <<0.001. (C) Mean clonogenic frequencies are consistent across donor proportions. Data are a mean of two independent cords with a total of 30 cells for each cord at each dose analysed except the 0% condition which only had 20 cells per cord. (D) Zygosity can be adjusted by inclusion of silent donor. Mean frequencies of homozygous mutant, heterozygous, and homozygous silent donors are shown within all clones with any observed editing. A total of 52 clones with some degree of editing across two independent cords were analyzed.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | AF647 (mouse monoclonal) Anti-Human CD34 (clone 581) | Cedarlane | 343508 | 1:200 |
| Antibody | V450 (mouse monoclonal) Anti-Human CD45RA (clone HI100) | BD Biosciences | 560362 | 1:100 |
| Antibody | PE-CF594 (mouse monoclonal) Anti-Human CD90 (clone 5E10) | BD Biosciences | 562385 | 1:200 |
| Antibody | FITC anti-human, CD49c (Clone REA360) | Miltenyi | 130-105-364 | 1:50 |
| Genetic reagent (AAV2/6) | Custom AAV6 – pssAAV_SRSF2P95H | Canadian Neurophotonics Platform – Viral Vector Core | Custom AAV | See annotated sequences in Supplementary file 2 |
| Genetic reagent (AAV2/6) | Custom AAV6 – pssAAV_SF3B1 K700E | Canadian Neurophotonics Platform – Viral Vector Core | Custom AAV | See annotated sequences in Supplementary file 2 |
| Biological sample (Homo sapiens) | Human Cord Blood for CD34 + cells harvest | Héma Québec via St Justine hospital | NA | |
| Cell line (Mus musculus) | M210B4 expressing human IL-3 and G-CSF | Gift from Connie J Eaves | NA | *special request |
| Cell line (Mus musculus) | sl/sl mouse fibroblasts expressin human SCF and IL-3 | Gift from Connie J Eaves | NA | *special request |
| Cell line (Mus musculus) | sl/sl mouse fibroblasts expressin human FLT3L | Gift from Connie J Eaves | NA | *special request |
| Chemical compound, drug | UM 171 | ExcellThera | NA | *special request |
| Chemical compound, drug | RS-1 | Cedarlane | 21037–5 | |
| Chemical compound, drug | Nedisertib (M3814) | Cedarlane | A17055 | |
| Chemical compound, drug | AZD 7648 | Cedarlane (Cayman) | 28598–1 | |
| Chemical compound, drug | DIMETHYL SULFOXIDE (DMSO), Sterile | BioShop | DMS666.100 | |
| Chemical compound, drug | Hydrocortisone | BioShop | HYD400.5 | |
| Chemical compound, drug | 1 M buffer | Homemade | Homemade | |
| Commercial assay, kit | PLATINUM SUPERFI II MASTER MIX | Life Technologies | 12368050 | |
| Commercial assay, kit | StemSpan CC100 | STEMCELL Technologies | 2690 | |
| Commercial assay, kit | MyeloCult H5100 | STEMCELL Technologies | 05150 | |
| Commercial assay, kit | Blunt/TA Ligase Master Mix | NEB | M0367S | |
| Commercial assay, kit | NEBNext Quick Ligation Module | NEB | E6056S | |
| Commercial assay, kit | NEBNext Ultra II End Repair/dA-Tailing Module | NEB | E7546S | |
| Commercial assay, kit | EasySep Human CD34 Positive Selection Kit II | STEMCELL Technologies | 17896 | |
| Commercial assay, kit | MethoCult H4034 Optimum | STEMCELL Technologies | 4034 | |
| Commercial assay, kit | Flongle Sequencing Expansion | Oxford Nanopore | FLO-FLG114 | |
| Commercial assay, kit | Flongle Flow Cell (R10.4.1) | Oxford Nanopore | FLO-FLG114 | |
| Commercial assay, kit | Native Barcoding Kit 24 V14 | Oxford Nanopore | SQK-NBD114.24 | |
| Commercial assay, kit | FBS Canadien | Thermo | 12483020 | |
| Commercial assay, kit | RPMI1640 | LifeTech | 11875119 | |
| Commercial assay, kit | StemSpan SFEM II | STEMCELL Technologies | 9655 | |
| Peptide, recombinant protein | Alt-R S.p. Cas9 Nuclease V3, 500 µg | IDT | 1081058 | |
| Peptide, recombinant protein | T7 Endonuclease I - 250 units | NEB | M0302S | |
| Peptide, recombinant protein | Proteinase K, Molecular Biology Grade | NEB | P8107S | |
| Peptide, recombinant protein | Alt-R Cas9 Electroporation Enhancer, 2 nmol | IDT | 1075915 | |
| Peptide, recombinant protein | BspEI Enzyme | NEB | R0540S | |
| Peptide, recombinant protein | IL-3, Human (CHO-expressed), 100 ng/ul | Cedarlane (GeneScript) | Z02991-10 | |
| Peptide, recombinant protein | SCF, Human (P. pastoris-expressed), 100 ng/ul | Cedarlane (GeneScript) | Z02692-10 | |
| Peptide, recombinant protein | EPO 100 ng/ul (~16 IU/uL) | Cedarlane (GeneScript) | Z02975-10 | |
| Peptide, recombinant protein | Flt-3L 100 ng/ul | Cedarlane (GeneScript) | Z02926-10 | |
| Peptide, recombinant protein | GM-CSF, Human (CHO-expressed), 100 ng/uL | Cedarlane (GeneScript) | Z02983-10 | |
| Peptide, recombinant protein | IL-6, Human (CHO-expressed), 100 ng/uL | Cedarlane (GeneScript) | Z03134-50 | |
| Peptide, recombinant protein | G-CSF, Human (CHO-expressed), 100 ng/uL | Cedarlane (GeneScript) | Z02980-10 | |
| Peptide, recombinant protein | CellAdhere Type I Collagen, Bovine, Solution | STEMCELL Technologies | 7001 | |
| Recombinant DNA reagent | p53 siRNA id s605 | Thermo | 4390824 | |
| Sequence-based reagent | SRSF2_gRNA1 | IDT | /AlTR1/rCrGrGrCrUrGrUrG rGrUrGrUrGrArGrUrCrCr GrGrGrUrUrUrUrArGrAr GrCrUrArUrGrCrU/AlTR2/ | crRNA SRSF2 |
| Sequence-based reagent | pri0077-F | IDT | AGCGATATAAACGGGCGCAG | Outer PCR SRSF2 |
| Sequence-based reagent | pri0077-R | IDT | TCGCGACCTGGATTTGGATT | Outer PCR SRSF2 |
| Sequence-based reagent | pri0002-H3 | IDT | CTATGGATGCCATGGACGGG | Inner PCR SRSF2 |
| Sequence-based reagent | pri0002-H4 | IDT | CAAGCACAGCGGGGTTAATTC | Inner PCR SRSF2 |
| Sequence-based reagent | pri0261-F | IDT | TCATTGGCAAACAGCAAGCC | SRSF2 gRNA1 off-target 1 |
| Sequence-based reagent | pri0261-R | IDT | AGAAGTATGTGCCTACGCGG | SRSF2 gRNA1 off-target 1 |
| Sequence-based reagent | pri0262-F | IDT | GAGAGTCACCGACCATGACG | SRSF2 gRNA1 off-target 2 |
| Sequence-based reagent | pri0262-R | IDT | TGTAAAACGTGCTGGAGGCT | SRSF2 gRNA1 off-target 2 |
| Sequence-based reagent | pri0263-F | IDT | CAGAAAGCACAAGCAACGCT | SRSF2 gRNA1 off-target 3 |
| Sequence-based reagent | pri0263-R | IDT | TCTCTTCCGGACACAAGTGC | SRSF2 gRNA1 off-target 3 |
| Sequence-based reagent | pri0285 | IDT | CTCCTTCTTCACGTCTTCCT | SRSF2 off-target 2 sequencing |
| Sequence-based reagent | pri0286 | IDT | CACCACATCTGGGATCCTCA | SRSF2 off-target 3 sequencing |
| Sequence-based reagent | SF3B1 Cas9 gRNA K700 | IDT | /AlTR1/rUrGrGrArUrGrArGrCrArGr CrArGrArArArGrUrUrGrUrUrUrUrAr GrArGrCrUrArUrGrCrU/AlTR2/ | crRNA SF3B1 |
| Sequence-based reagent | Alt-R CRISPR-Cas9 tracrRNA | IDT | 1072533 | tracrRNA |
| Sequence-based reagent | pri0078-F | IDT | GCTGCTGGTCTGGCTACTAT | Outer PCR SF3B1 |
| Sequence-based reagent | pri0078-R | IDT | ATACTCATTGCTGATTACGTGATTT | Outer PCR SF3B1 |
| Sequence-based reagent | pri0002-H1 | IDT | TGGGCTACTGATTTGGGGAG | Inner PCR SF3B1 |
| Sequence-based reagent | pri0002-H2 | IDT | CTGTGTTGGCGGATACCCTT | Inner PCR SF3B1 |
| Sequence-based reagent | SRSF2 Silent ssODN | IDT | t*g*gacggccgcgagctgcgggtgcaaatg gcgcgctacggccgcccTccAgaTtcacacca cagccgccggggaccgccaccccgcag*g*t | |
| Sequence-based reagent | SRSF2 P95H ssODN | IDT | t*g*gacggccgcgagctgcgggtgcaaatggcg cgctacggccgccATccggactcacaccacag ccgccggggaccgccaccccgcag*g*t | |
| Sequence-based reagent | SRSF2 long ssODN donor | IDT | Ttcacgacaagcgcgacgctgaggacgctatgga Tgccatggacggggccgtgctggacggccgcga gctgcgggtgcaaatggcgcgctacgg ccgccATccggactcacaccacagccgccggg gaccgccaccccgcaggtacgggggcggtggcta cggacgccggagccgcaggtaaacgg ggctgaggggaccg | ordered as ALT-R HDR Donor Oligo |
| Sequence-based reagent | SRSF2 P95H ssODN with additional silent mutations | IDT | t*g*gacggccgcgagctgcgggtgcaaatggcgcgctac ggccgccATccAgaTtcacaccacagccgccggg gaccgccaccccgcag*g*t | |
| Software and algorithms | GelAnalyzer 19.1 | Istvan Lazar Jr. and Istvan Lazar Sr. | https://www.gelanalyzer.com | |
| Software and algorithms | Synthego Performance Analysis V3 | ICE Analysis | https://www.synthego.com | |
| Software and algorithms | FlowJo Software 10.8.1 | BD Life Sciences | https://www.flowjo.com | |
| Software and algorithms | R (version 4.1.2) | R Core Team | https://www.r-project.org/ | |
| Software and algorithms | MinKNOW (version 23.11.3) | Oxford Nanopore | https://community.nanoporetech.com/downloads | |
| Software and algorithms | minimap2 (version 2.26) | Dana-Farber Cancer Institute | https://github.com/lh3/minimap2 | |
| Software and algorithms | samtools (version 1.10) | Genome Research Ltd. | http://www.htslib.org/ | |
| Software and algorithms | bcftools (version 1.10.2) | Genome Research Ltd. | https://samtools.github.io/bcftools/ | |
| Software and algorithms | Biopython (version 1.83) | Biopython | https://biopython.org/ | |
| Software and algorithms | Python (version 3.9.7) | Python | https://www.python.org/ |
Additional files
-
Supplementary file 1
Significance testing.
Figure and panel are indicated along with each pair-wise test. Where relevant the colony type or population for a given test is indicated.
- https://cdn.elifesciences.org/articles/91288/elife-91288-supp1-v1.docx
-
Supplementary file 2
Annotated DNA sequence used in the experiments for HDR testing and integration.
- https://cdn.elifesciences.org/articles/91288/elife-91288-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91288/elife-91288-mdarchecklist1-v1.docx
-
Source data 1
Raw gel images.
Data file for the raw gels compiled in Source data 2.
- https://cdn.elifesciences.org/articles/91288/elife-91288-data1-v1.zip
-
Source data 2
Annotated gels.
Compilation of the annotated gels used for quantification of RNP and HDR integration efficiencies.
- https://cdn.elifesciences.org/articles/91288/elife-91288-data2-v1.pdf
-
Source data 3
Numeric data.
Compilation of the numeric data used throughout the manuscript.
- https://cdn.elifesciences.org/articles/91288/elife-91288-data3-v1.zip





