Mono-methylated histones control PARP-1 in chromatin and transcription
Figures
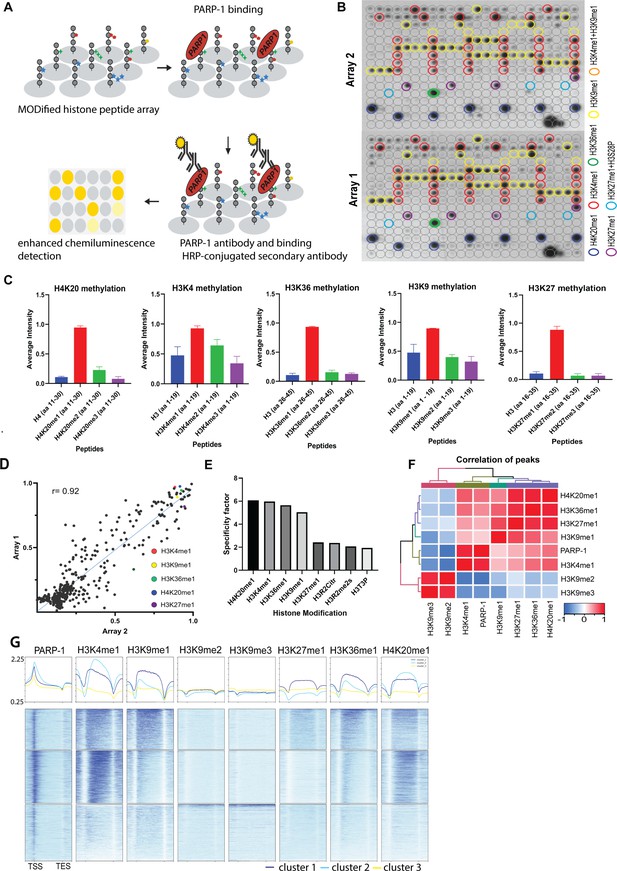
PARP-1 binds H4K20me1, H3K4me1, H3K36me1, H3K9me1, and H3K27me1 in vitro and in vivo.
(A) Illustration of the MODified histone peptide assay (Active Motif) used to determine PARP-1 binding to histone modifications. The histone peptide array (Top, left), comprising 19mer peptides with single or up to four concurrent histone modifications, was employed to investigate PARP-1’s binding affinity for histone modifications and to assess the impact of adjacent modified peptides on PARP-1 binding. This array was first blocked, then incubated with PARP-1 protein (Top, right). Subsequently, it was stained with a PARP-1 antibody and a horseradish peroxidase (HRP)-conjugated secondary antibody (Bottom, right). Visualization of PARP-1 binding was done through enhanced chemiluminescence detection and captured on X-ray film (Bottom, left). See Methods for a full description. (B) Signal intensity on modified histone peptide array based on incubation with PARP-1 protein. (C) Average intensities of PARP-1 binding to single histone peptides. Data are presented as mean ± SEM (D–E) Reproducibility and specificity of spot intensities from modified histone peptide array duplicates. (D) Scatter plot showing the correlation of the average intensities of duplicate arrays. Intensities of PARP-1 binding to all peptides and spots containing single H4K20me1, H3K4me1, H3K36me1, H3K9me1, and H3K27me1 (key) are shown. Pearson’s correlation coefficient (r) is 0.92. (E) Bar chart showing top 8 histone modifications with the highest specificity for PARP-1 binding. The specificity factor was calculated by dividing the average intensity of spots that contain the modified histone peptide by the average intensity of spots that do not contain the peptide. (F) Spearman correlation of PARP-1, H4K20me1, H3K4me1, H3K36me1, H3K9me1, H3K9me2/3, and H3K27me1 peaks in Drosophila third-instar larvae based on fraction of overlap. (G) Heatmaps showing k-means clustering-generated occupancy of PARP-1, H4K20me1, H3K4me1, H3K36me1, H3K9me1, H3K9me2/3, and H3K27me1 normalized ChIP-seq signals in third-instar larvae at Drosophila genes. ChIP-seq signals are sorted in descending order. The upper plots show the summary of the signals.
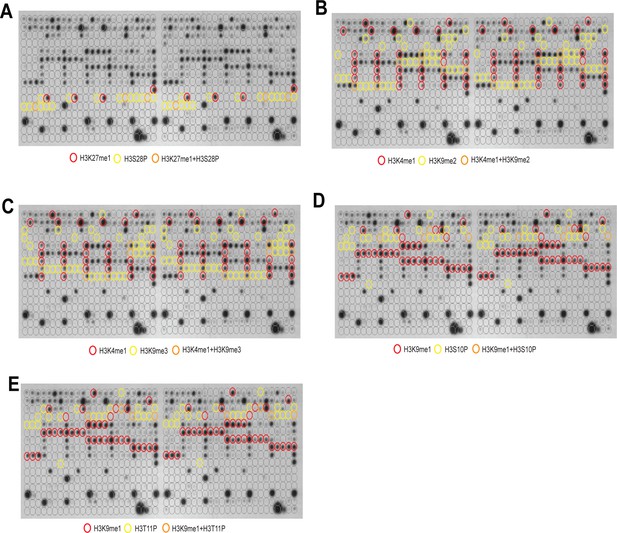
PARP-1 binding is inhibited by H3K9me2/3 peptides or by phosphorylation of adjacent residues.
Signal intensity on modified histone peptide array showing inhibition of PARP-1 binding to spots containing H3K27me1 by the presence of H3S28P peptides (A), inhibition of PARP-1 binding to spots containing H3K4me1 peptides by the presence of H3K9me2 (B) or H3K9me3 (C) peptides, inhibition of PARP-1 binding to spots containing H3K9me1 peptides by the presence of H3S10P (D) or H3T11P (E) peptides. Blots on duplicate arrays are shown.
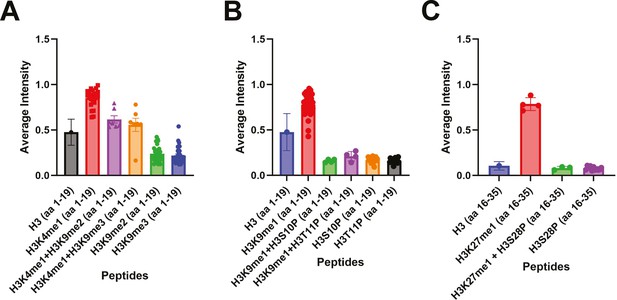
Inhibition of PARP-1 binding by H3K9me2/3 and phosphorylation of adjacent residues.
Average intensities from histone peptide array showing the inhibition of PARP-1 binding to spots containing H3K4me1 peptides by the presence of H3K9me2/3 peptide (A), inhibition of PARP-1 binding to spots containing H3K9me1 peptides by the presence of H3S10P or H3T11P peptides (B), inhibition of PARP-1 binding to spots containing H3K27me1 peptides by the presence of H3S28P peptides (C).
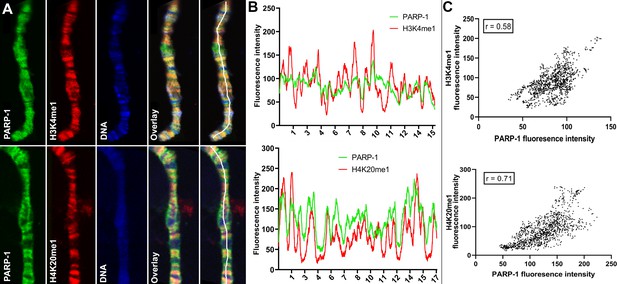
PARP-1 colocalizes with H3K4me1 and H4K20me1 histone marks in polytene chromosomes.
(A) Immunofluorescent staining of Drosophila salivary gland chromosomes showing colocalization of PARP-1 with H3K4me1 and H4K20me1. White lines indicate areas of the colocalization-quantification shown in (B). (B–C) Quantification of fluorescence intensity of PARP-1, H3K4me1, and H4K20me1 at Drosophila polytene chromosomes in panel A. (B) Images show the distribution of PARP-1 fluorescence Intensity with H3K4me1 (top) and H4K20me1 (bottom) fluorescence intensity. (C) Images represent a scatterplot showing PARP-1 fluorescence intensity with H3K4me1 (top) and H4K20me1 (bottom) fluorescence intensity. Pearson correlation coefficients (r) are respectively 0.58 (top) and 0.71 (bottom).
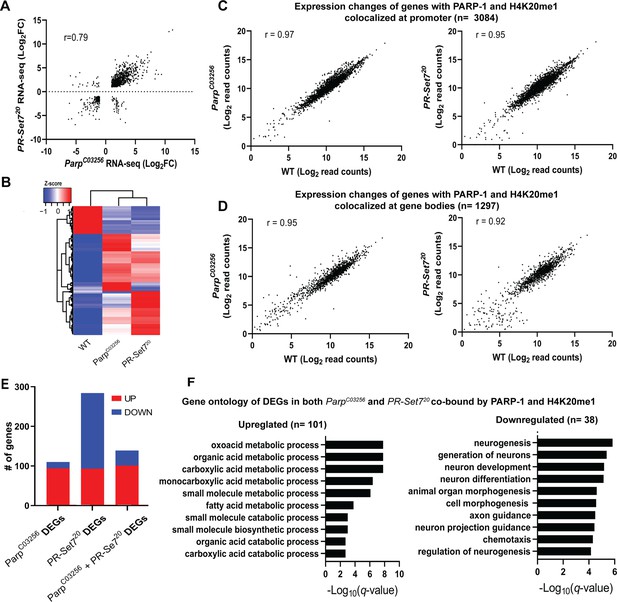
PARP-1 and H4K20me1 are required for the repression of metabolic genes and activation of developmental genes at co-enriched genes.
(A) Scatterplot plot showing correlation of differentially expressed genes (DEGs) in parp-1C03256 and pr-set720 (Pearson’s r=0.79). (B) Heatmap showing the normalized read counts of DEGs in both parp-1C03256 and pr-set720. Normalized read counts are shown as row z-scores. (C–D) Dot plots showing transcriptional changes of genes co-enriched with PARP-1 and H4K20me1 in parp-1C03256 and pr-set720 compared to WT at promoters (C) and gene bodies (D). (E) Summary of DEGs in parp-1C03256 and pr-set720 and both mutants that were co-enriched with PARP-1 and H4K20me1. (F) Gene ontology of upregulated (left) and downregulated (right) DEGs in both parp-1C03256 and pr-set720 mutants that were co-enriched with PARP-1 and H4K20me1.
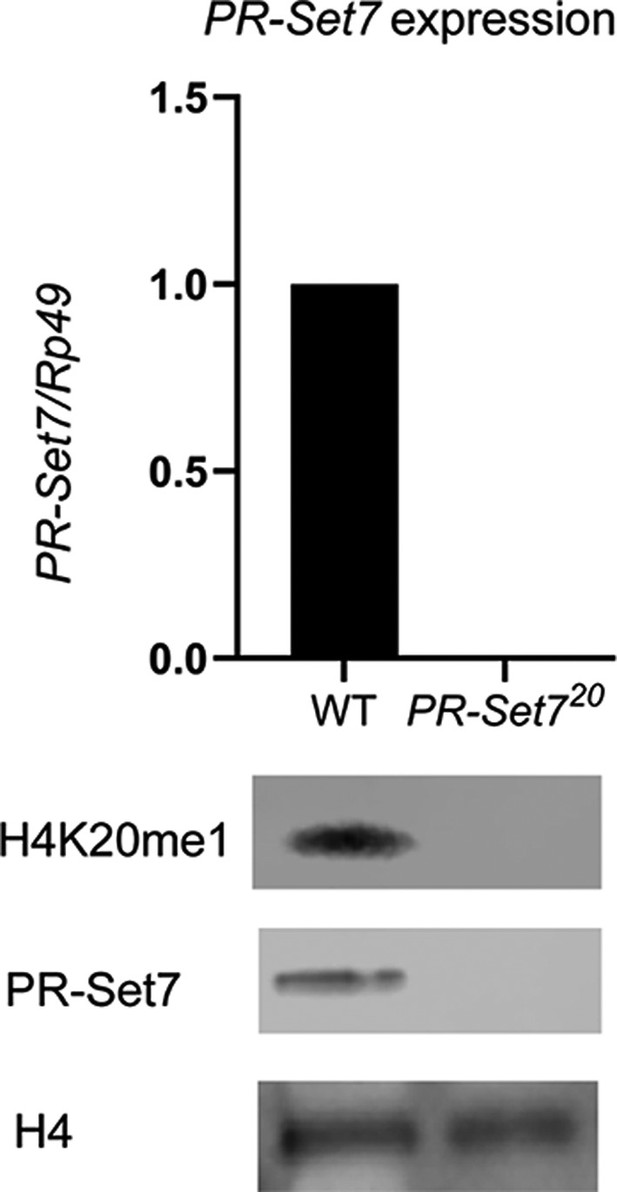
Validation of pr-set720 mutant.
(Top) Expression levels of PR-SET7 in wild-type (WT) or pr-set720 mutant during third-instar larval stage (three biological replicates). (Bottom) Western blot of H4K20me1, PR-SET7, and H4 loading control in WT or pr-set720 mutant during third-instar larval stage (two biological replicates).
-
Figure 3—figure supplement 1—source data 1
Original file for the Western blot analysis in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/91482/elife-91482-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Figure 3—figure supplement 1 and original scans of the relevant Western blot analysis with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/91482/elife-91482-fig3-figsupp1-data2-v1.zip
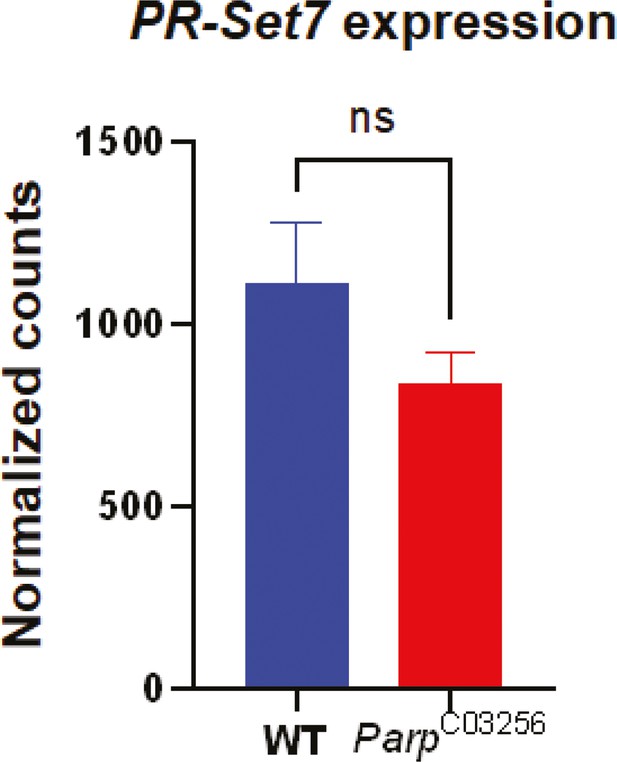
PR-SET7 expression level is not affected in parp-1C03256 mutant.
Normalized counts of Pr-SET7 RNA in a wild-type (blue) or in a parp-1C03256 (red) background based on RNA-seq data (GSE222877). The experiment was performed in triplicates. The statistical test is an unpaired two-tailed t-test. (ns): non-significant. Data are presented as mean ± SEM.
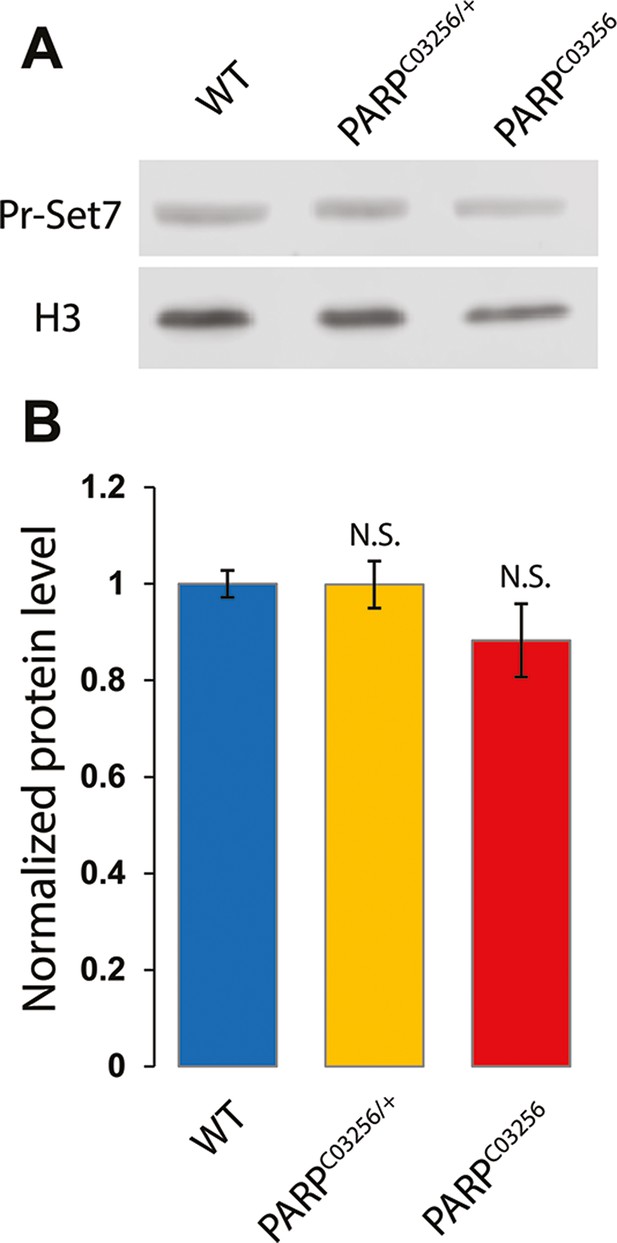
PR-SET7 protein level is not affected in parp-1C03256 mutant.
(A) Western blot showing Pr-SET7 protein level in a wild-type (WT), parp-1 heterozygote (ParpC03256/+), or homozygote mutant (ParpC03256) background. H3 is used as a loading control. (B) Quantification of PR-SET7 protein level based on three independent biological replicates and normalized to H3 level. The statistical test is an unpaired two-tailed t-test. N.S: non-significant.
-
Figure 3—figure supplement 3—source data 1
Original file for the Western blot analysis in Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/91482/elife-91482-fig3-figsupp3-data1-v1.zip
-
Figure 3—figure supplement 3—source data 2
This file containing Figure 3—figure supplement 3 and original scans of the relevant Western blot analysis with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/91482/elife-91482-fig3-figsupp3-data2-v1.zip
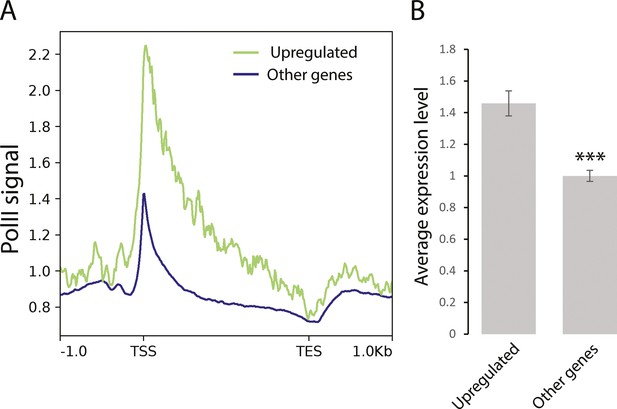
Genes coenriched in H4K20me1 and PARP-1 that are upregulated in both parp-1 and pr-set7 mutants are highly expressed genes.
(A) Distribution of RNA-Polymerase II (PolII) protein along genes that are coenriched by H4K20me1 and PARP-1 and upregulated both parp-1 and pr-set7 mutants (green) and along all other Drosophila genes (blue). The region of all target genes is scaled to a 2 kb region (from TSS to TES) and includes the 1 kb region upstream from transcription start site (TSS) and downstream from TES. The analysis was performed on the whole organism on Drosophila third instar larval puffstage 7–9. (B) Average expression level of genes coenriched by H4K20me1 and PARP-1 and upregulated both parp-1 and pr-set7 mutants (upregulated) and average expression of all other genes. The expression was measured on the whole organism on Drosophila third instar larval puffstage 7–9. Statistical test is an unpaired two-tailed t-test. ***p-value <0.01.
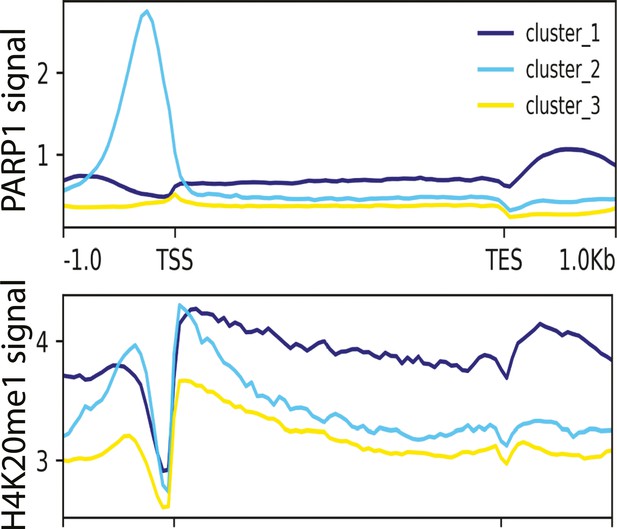
PARP-1 binding correlates with H4K20me1 enrichment in Human K562 cells.
Metagene plots showing enrichment of PARP-1 (CUT&Tag) and H4K20me1 (ChIP-Seq) at PARP-1 clusters (cluster 1=5385, cluster 2=2637, cluster 3=11, 920 regions) in human protein-coding genes. Signals extend from −1 kb of the TSS to +1 kb of the TES.

Dynamic H4K20me1 enrichment regulates the expression of heat-shock genes during heat shock.
(A) Expression of hsp22, hsp23, hsp68, hsp70, and hsp83 in wild-type (WT), parp-1C03256 and pr-set720 third-instar larvae before and after 30 min heat shock. Data shown are from three to five biological replicates. ***p<0.001, **p<0.01, *p<0.05 (Unpaired t-test; Two-tailed). Integrative Genome Viewer (IGV) tracks showing normalized ChIP-seq tracks of PARP-1 and H4K20me1 in third-instar larvae at (B) hsp22, hsp68, hsp68, hsp70, and (C) hsp22, hsp83 before heat shock. Mononucleosome ChIP-qPCR in WT showing enrichment of H4K20me1 at (D) hsp70, hsp22, hsp68, and (E) hsp23 and hsp83 before heat shock (NHS), after 15 min heat shock and 30 min heat shock. Primers used spanned the hsp70 locus and the gene bodies of hsp22, hsp23, hsp68, hsp70, and hsp83. Data shown are from three biological replicates. Data are presented as mean ± SEM.
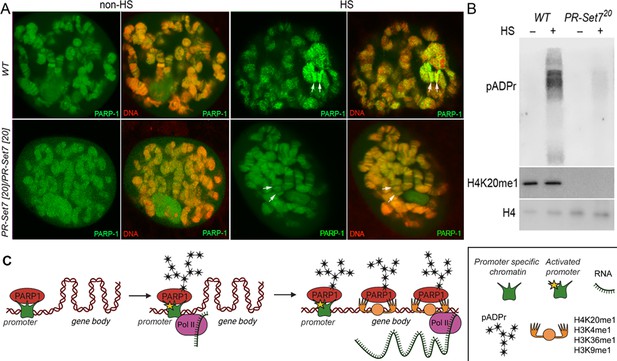
Mono-methylated histones controls PARP-1 binding along gene body to regulate transcription.
(A) PR-SET7/H4K20me1 is required for holding PARP-1 in chromatin during heat shock (HS). PARP-1 (green) protein recruitment to hsp70 locus (Arrows) in salivary gland polytene nuclei in wild-type (WT) and pr-set720 mutant third instar larvae. A single salivary gland polytenized nucleus of wandering third instar larvae is shown for each genotype. Wild-type genotype - UASt::PARP-1-EYFP, GAL4[Mz1087.hx]; pr-set7 mutant genotype – UASt::PARP-1-EYFP, GAL4Mz1087.hx; pr-set720. (B) Equal amounts of lysates from the wild-type expressing PARP-1-EYFP (WT) and pr-set720 mutants expressing PARP-1-EYFP (pr-set720) grown at 22 °C or heat shocked at 37 °C for 1 hr at the third-instar larvae stage were subjected to immunoblot analysis using mouse anti-pADPr (10 H), anti-H4K20me1 and anti-histone H4 (loading control) antibodies. (C) Model of PARP-1 regulation by histone modifications. PARP-1 binds to a nucleosome that carries the H2A variant (H2Av) at the promoter region. Upon developmental triggers or heat shock-induced phosphorylation of H2Av, PARP-1 is activated (Kotova et al., 2011). Activated PARP-1 fosters a transcription start site (TSS) that is more accessible, thereby enabling the binding of RNA-Polymerase II (Pol II) and initiation of transcription (Nikolaou et al., 2017). Following this, the distribution of PARP-1 is further enhanced by active mono-methylated forms of H4K20, H3K4, H3K36, or H3K9. Each of these histone modifications can interchangeably facilitate this function. The spreading of PARP-1 to the gene body contributes to the loosening of chromatin in this region (Tulin and Spradling, 2003; Estève et al., 2022; Petesch and Lis, 2012; Rickels et al., 2017). Consequently, this facilitates the transition of Pol II into a productive elongation phase, leading to the generation of a mature transcript.
-
Figure 5—source data 1
Original file for the Western blot analysis in Figure 5B.
- https://cdn.elifesciences.org/articles/91482/elife-91482-fig5-data1-v1.zip
-
Figure 5—source data 2
This file contains Figure 5B and original scans of the relevant Western blot analysis with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/91482/elife-91482-fig5-data2-v1.zip
Additional files
-
Supplementary file 1
Data of the PARP-1 histone peptide array.
Sheet 1 displays raw and processed data for each modification tested. Column 1 displays the peptide number. Column B displays the location on the plate. Column C displays the name of the Histone tail. Columns D to G displays the modification(s) present on the histone tail. Columns H and I display the intensity recorded for the peptide in Array 1 and 2, respectively. Column J displays the average intensity, calculated based on values from columns H and I. Column K displays the standard error of mean based on values from columns H and I. Sheet 2 displays the specificity factor for each histone modification. Column A displays the rank of the modification (based on their specificity factor). Column B displays the name of the histone modification. Columns C and D display the number of dots of the array that contain the modification (column C) or that does not contain the modification (column D). Columns E and F display the average intensity of the positive (E) or negative (F) dots. Column G displays the specificity factor of the histone modification.
- https://cdn.elifesciences.org/articles/91482/elife-91482-supp1-v1.xlsx
-
Supplementary file 2
Primers used in this study.
This supplementary file displays the list of primers used for qPCR (sheet 1) or for ChIP-qPCR (sheet 2) in this study.
- https://cdn.elifesciences.org/articles/91482/elife-91482-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91482/elife-91482-mdarchecklist1-v1.docx





