Xanthomonas citri subsp. citri type III effector PthA4 directs the dynamical expression of a putative citrus carbohydrate-binding protein gene for canker formation
Figures
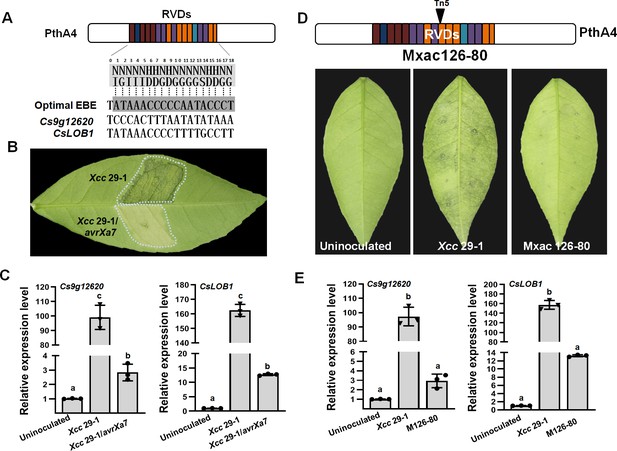
The expression of Cs9g12620 depends on PthA4 during Xcc infection.
(A) Predicted PthA4-binding elements of the Citrus sinensis genes upregulated in response to Xcc infection. The optimal effector-binding element (EBE) that corresponded to the PthA4 repeat variable di-residues (RVDs) were predicted based on the transcriptional activator-like (TAL) effector–DNA-binding code. Putative PthA4 EBEs in promoters of Cs9g12620 and CsLOB1 are shown below the optimal EBE of PthA4. (B) The disease symptoms of Xcc 29-1/avrXa7 on C. sinensis leaves. The cell suspension (108 CFU/ml) was infiltrated into plant leaves, and the phenotype was recorded at 5 dpi. All the experiments were repeated three times with similar results. (C) A qRT-PCR analysis of the expression of Cs9g12620 and CsLOB1 in plants inoculated with Xcc that expressed avrXa7. The level of expression of each gene in the uninoculated samples was set as 1, and the levels in other samples were calculated relative to those baseline values. Values are the mean results from three biological replicates and are the means ± SD (ANOVA, p < 0.01). The experiment was repeated three times. (D) The phenotype elicited by the pthA4 mutant Mxac126-80. The analyses were performed as described in B. (E) qRT-PCR analysis of the transcript levels of Cs9g12620 and CsLOB1 plants inoculated with Xcc 29-1 and Mxac126-80. The analyses were the same as those conducted in C. ANOVA, analysis of variance; dpi, days post-inoculation; qRT-PCR, real-time quantitative reverse transcription PCR; SD, standard deviation; Xcc, Xanthomonas citri subsp. citri.
-
Figure 1—source data 1
Excel file containing qRT raw data in Figure 1C, E.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Original image for the disease symptoms analysis in Figure 1B (Xcc 29-1, Xcc 29-1/avrXa7).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig1-data2-v1.zip
-
Figure 1—source data 3
PDF containing Figure 1B and original scans of the disease symptoms analysis (Xcc 29-1, Xcc 29-1/avrXa7) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig1-data3-v1.pdf
-
Figure 1—source data 4
Original image for the disease symptoms analysis in Figure 1D (uninoculated).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig1-data4-v1.zip
-
Figure 1—source data 5
Original image for the disease symptoms analysis in Figure 1D (Xcc 29-1).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig1-data5-v1.zip
-
Figure 1—source data 6
Original image for the disease symptoms analysis in Figure 1D (Mxac126-80).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig1-data6-v1.zip

The location of the PthA4 effector-binding element (EBE) in the promoter of Cs9g12620 in Citrus sinensis.
The promoter sequences were retrieved from the citrus genome database. The location of binding site of PthA4 EBE is indicated by an underline. Translation initiation site is indicated by an arrow.
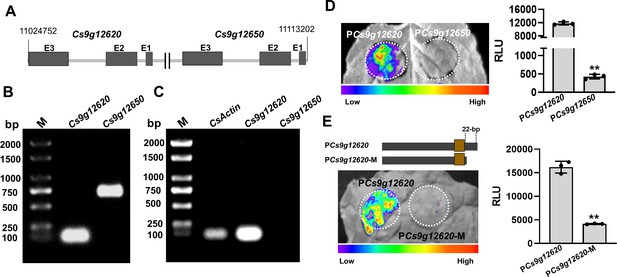
Cs9g12620 and Cs9g12650 show different profiles of expression owing to the genetic variation in promoter.
(A) The location of Cs9g12620 and Cs9g12650 in the Citrus sinensis chromosome. The location was revealed by a BLAST search of the Citrus Genome Database (https://www.citrusgenomedb.org/). (B) Verification of the existence of Cs9g12620 and Cs9g12650 in the C. sinensis genome by PCR amplification. The specific primers for either gene were used for qRT-PCR analysis, which are shown in Supplementary file 1b. M, DL2000 DNA marker. (C) Semi-quantitative RT-PCR detection of the transcription of Cs9g12620 and Cs9g12650. CsActin was used as the internal control gene. A total of 2 µg of total RNA extracted from C. sinensis was used to synthesize the first single-stranded cDNA. The specific primers for each gene were the same as those of the qRT-PCR analysis and are shown in Supplementary file 1b. M, DL2000 DNA marker. (D) Luciferase assays of Cs9g12620 and Cs9g12650 promoter activity. The Cs9g12620 and Cs9g12650 promoter luciferase fusions were transiently expressed in the Nicotiana benthamiana leaves. Luciferase activity was measured with a CCD imaging system at 2 days post-agroinfiltration. The image on the right shows the quantification of luciferase signal using a microplate luminescence reader. Values are the means ± SD (n = 3 biological replicates; Student’s t-test, **p ˂ 0.01). (E) Luciferase assays of Cs9g12620 and Cs9g12620-M with the deletion of 22 bp at the 3′-terminus. The Cs9g12620 and Cs9g12620-M promoter luciferase fusions were transiently expressed in the N. benthamiana leaves. Luciferase activity was measured with a CCD imaging system at 2 days post-agroinfiltration. The luciferase signal was quantified as described in D. CCD, charge-coupling device; qRT-PCR, real-time quantitative reverse transcription PCR; RT-PCR, reverse transcription PCR; SD, standard deviation.
-
Figure 2—source data 1
Excel file containing RLU raw data in Figure 2D, E.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Original file for the PCR amplification analysis in Figure 2B (Cs9g12620 and Cs9g12650).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data2-v1.zip
-
Figure 2—source data 3
PDF containing Figure 2B and original scans of the PCR amplification analysis (Cs9g12620 and Cs9g12650) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data3-v1.pdf
-
Figure 2—source data 4
Original file for the semi-quantitative reverse transcription PCR (RT-PCR) analysis in Figure 2C (CsActin, Cs9g12620, and Cs9g12650).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data4-v1.zip
-
Figure 2—source data 5
PDF containing Figure 2C and original scans of the semi-quantitative reverse transcription PCR (RT-PCR) analysis (CsActin, Cs9g12620, and Cs9g12650) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data5-v1.pdf
-
Figure 2—source data 6
Original file for the luciferase assays analysis in Figure 2D (Cs9g12620 and Cs9g12650 promoter).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data6-v1.zip
-
Figure 2—source data 7
PDF containing Figure 2D and original scans of the luciferase assays analysis (Cs9g12620 and Cs9g12650 promoter) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data7-v1.pdf
-
Figure 2—source data 8
Original file for the luciferase assays analysis in Figure 2E (Cs9g12620 and Cs9g12620-M promoter).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data8-v1.zip
-
Figure 2—source data 9
PDF containing Figure 2E and original scans of the luciferase assays analysis (Cs9g12620 and Cs9g12620-M promoter) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig2-data9-v1.pdf

Alignment of the promoter regions of Cs9g12620 and Cs9g12650 in Citrus sinensis.
The promoter sequences were retrieved from the citrus genome database. The putative binding sites of CsLOB1 (LB1 and LB2) are shadowed with blue. The location of binding site of PthA4 (effector-binding element, EBE) is indicated by an underline. A green shadow shows the location of predicted promoter core structure. The lack of 22 bp at the 3′ terminus is shown by a red box.
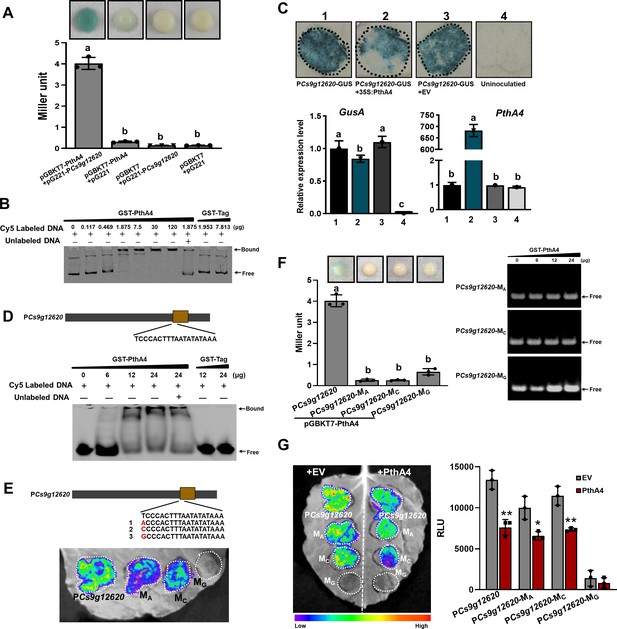
PthA4 binds to and suppresses the Cs9g12620 promoter.
(A) A yeast one-hybrid assay shows that PthA4 interacts with the Cs9g12620 promoter. pGBKT7-pthA4 and pG221-PCs9g12620 were co-transformed into the yeast strain EGY48 and screened on synthetic dropout SD/-Ura and SD/-Ura/-Trp media. The transformed cells were dissociated by repeated freeze–thaw treatment and then used for β-galactosidase assays on filters soaked with Z buffer that contained 20 μg/ml X-gal. The pGBKT7 and pG221 vectors were used as negative controls by co-transformation with the corresponding constructs. The columns below show the quantitative assay of β-galactosidase by the Miller method. Values are the means ± SD (n = 3 biological replicates). Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (B) An electrophoretic mobility shift assay (EMSA) shows that PthA4 binds to the Cs9g12620 promoter. Purified GST-PthA4 was incubated with 25 ng of promoter DNA in gel shift binding buffer at 28°C for 30 min and then analyzed in 6% non-denaturing PAGE. A GST tag was used as a negative control. (C) The overexpression of PthA4 suppressed theCs9g12620 promoter. PthA4 was transiently co-expressed with a Cs9g12620 promoter GUS fusion in Nicotiana benthamiana. The promoter-driven GUS activity was assayed at 2 days post-agroinfiltration. The upper images show histochemical staining of the Cs9g12620 promoter-driven GUS activity. The bar chart below shows the qRT-PCR analysis of the transcript levels of gusA and pthA4. The level of expression of each gene in the samples that expressed the promoter alone was set as 1, and the levels of the other samples were calculated relative to that. Values are the mean results from three biological replicates and are the means ± SD. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (D) EMSA shows that PthA4 binds to the 19 bp predicted binding site (effector-binding element, EBE) in the Cs9g12620 promoter. The purified GST-PthA4 was incubated with 25 ng of synthetic 19 bp EBE DNA fragment in gel shift binding buffer at 28°C for 30 min and then analyzed in 6% non-denaturing PAGE. (E) Luciferase assays of the activity of Cs9g12620 promoter with site-directed mutation in PthA4 EBE. The first nucleotide acid ‘T’ in EBE was mutated into A, C, or G, which generated PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG, respectively. The mutants were fused with luciferase and transiently overexpressed in N. benthamiana leaves. The luciferase activity was measured with a CCD imaging system at 2 days post-agroinfiltration. (F) PthA4 did not bind to PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG. The left image represents a yeast one-hybrid assay that did not show an interaction of PthA4 between PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG. The experiments were conducted in the same manner as those in A. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). The right image represents an EMSA that shows that PthA4 does not bind to the promoter mutants. Purified GST-PthA4 was incubated with 25 ng of promoter DNA in gel shift binding buffer at 28°C for 30 min and then analyzed in 6% non-denaturing PAGE. (G) Luciferase assays showing the suppression of PthA4 on PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG. The promoter luciferase fusions were co-expressed with PthA4 in N. benthamiana. Co-expression with the empty binary vector pHB was used as the control. The analysis was the same as that described in E. The image on the right shows the quantification of luciferase signal using a microplate luminescence reader. Values are the means ± SD (n = 3 biological replicates). ANOVA, analysis of variance; CCD, charge-coupling device; GST, glutathione-S transferase; PAGE, polyacrylamide gel electrophoresis; qRT-PCR, real-time quantitative reverse transcription PCR; SD, standard deviation. *p ˂ 0.05. **p ˂ 0.01 (Student’s t-test).
-
Figure 3—source data 1
Excel file containing LacZ, qRT and RLU raw data in Figure 3A, C, F, G.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Original file for the yeast one-hybrid assay in Figure 3A (pGBKT7-PthA4+pG221-PCs9g12620, pGBKT7+pG221-PCs9g12620, and pGBKT7+pG221).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data2-v1.zip
-
Figure 3—source data 3
Original file for the yeast one-hybrid assay in Figure 3A (pGBKT7-PthA4+pG221).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data3-v1.zip
-
Figure 3—source data 4
PDF containing Figure 3A and original scans of the yeast one-hybrid assay (pGBKT7-PthA4+pG221-PCs9g12620, pGBKT7+pG221-PCs9g12620, pGBKT7+pG221, and pGBKT7-PthA4+pG221) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data4-v1.pdf
-
Figure 3—source data 5
Original file for the electrophoretic mobility shift assay in Figure 3B (GST-PthA4+PCs9g12620).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data5-v1.zip
-
Figure 3—source data 6
PDF containing Figure 3B and original scans of the electrophoretic mobility shift assay (GST-PthA4+PCs9g12620) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data6-v1.pdf
-
Figure 3—source data 7
Original file for histochemical staining of the Cs9g12620 promoter-driven GUS activity in Figure 3C (PCs9g12620-GUS, PCs9g12620-GUS+35S:PthA4, PCs9g12620-GUS+EV, and uninoculatied).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data7-v1.zip
-
Figure 3—source data 8
PDF containing Figure 3C and original scans of the histochemical staining of the Cs9g12620 promoter-driven GUS activity assay (PCs9g12620-GUS, PCs9g12620-GUS+35S:PthA4, PCs9g12620-GUS+EV, and uninoculatied) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data8-v1.pdf
-
Figure 3—source data 9
Original file for the electrophoretic mobility shift assay in Figure 3D (GST-PthA4+19 bp effector-binding element [EBE] PCs9g12620).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data9-v1.zip
-
Figure 3—source data 10
PDF containing Figure 3D and original scans of the electrophoretic mobility shift assay (GST-PthA4+19 bp effector-binding element [EBE] PCs9g12620) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data10-v1.pdf
-
Figure 3—source data 11
Original file for the luciferase assays in Figure 3E (PCs9g12620, PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data11-v1.zip
-
Figure 3—source data 12
PDF containing Figure 3E and original scans of the luciferase assays (PCs9g12620, PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data12-v1.pdf
-
Figure 3—source data 13
Original file for the yeast one-hybrid assay in Figure 3F (pGBKT7-PthA4+pG221-PCs9g12620).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data13-v1.zip
-
Figure 3—source data 14
Original file for the yeast one-hybrid assay in Figure 3F (pGBKT7-PthA4+pG221-PCs9g12620-MA, pGBKT7-PthA4+pG221-PCs9g12620-MC, and pGBKT7-PthA4+pG221-PCs9g12620-MG).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data14-v1.zip
-
Figure 3—source data 15
PDF containing Figure 3F and original scans of the yeast one-hybrid assay (pGBKT7-PthA4+pG221-PCs9g12620, pGBKT7-PthA4+pG221-PCs9g12620-MA, pGBKT7-PthA4+pG221-PCs9g12620-MC, and pGBKT7-PthA4+pG221-PCs9g12620-MG) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data15-v1.pdf
-
Figure 3—source data 16
Original file for the electrophoretic mobility shift assay in Figure 3F (GST-PthA4+PCs9g12620-MA and GST-PthA4+PCs9g12620-MC).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data16-v1.zip
-
Figure 3—source data 17
Original file for the electrophoretic mobility shift assay in Figure 3F (GST-PthA4+PCs9 g12620-MG).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data17-v1.zip
-
Figure 3—source data 18
PDF containing Figure 3F and original scans of the electrophoretic mobility shift assay (GST-PthA4+PCs9g12620-MA, GST-PthA4+PCs9g12620-MC, and GST-PthA4+PCs9 g12620-MG) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data18-v1.pdf
-
Figure 3—source data 19
Original file for the luciferase assays in Figure 3G (PCs9g12620, PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG co-expression with PthA4 and EV, respectively).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data19-v1.zip
-
Figure 3—source data 20
PDF containing Figure 3G and original scans of the luciferase assays (PCs9g12620, PCs9g12620-MA, PCs9g12620-MC, and PCs9g12620-MG co-expression with PthA4 and EV, respectively) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig3-data20-v1.pdf
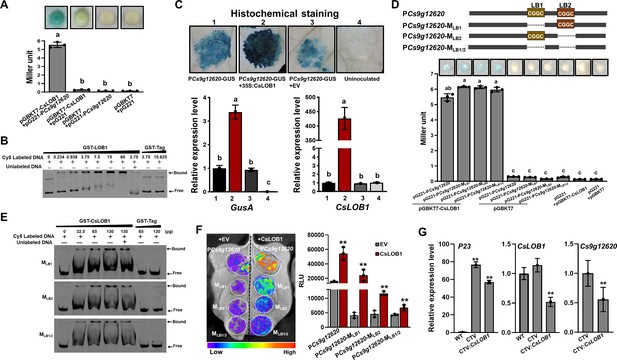
The Cs9g12620 promoter is directly activated by CsLOB1.
(A) A yeast one-hybrid (Y1H) assay shows that CsLOB1 interacts with the Cs9g12620 promoter. pGBKT7-CsLOB1 and pG221-PCs9g12620 were co-transformed into yeast strain EGY48 and screened on synthetic dropout SD/-Ura and SD/-Ura/-Trp media. The transformed cells were dissociated by repeated freeze–thaw treatment and then used for β-galactosidase assays on filters soaked with Z buffer that contained 20 μg/ml of X-gal. pGBKT7 and pG221 vectors were used as the negative control by co-transformation with corresponding constructs. The columns below show the quantitative assay of β-galactosidase by the Miller method. Values are the means ± SD (n = 3 biological replicates). Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (B) An electrophoretic mobility shift assay (EMSA) shows that CsLOB1 binds to the Cs9g12620 promoter. Purified GST-CsLOB1 was incubated with 25 ng of promoter DNA in gel shift binding buffer at 28°C for 30 min and was then analyzed by 6% non-denaturing PAGE. (C) Cs9g12620 promoter activity is induced by CsLOB1. CsLOB1 was transiently co-expressed with the Cs9g12620 promoter GUS fusion in Nicotiana benthamiana. Promoter-driven GUS activity was assayed at 2 days post-agroinfiltration. The upper images show the histochemical staining ofCs9g12620 promoter-driven GUS activity. The bar chart below shows the qRT-PCR analysis of transcript levels of gusA and pthA4. The level of expression of each gene in the samples that expressed the promoter alone was set as 1, and the levels of the other samples were calculated relative to that. Values are the mean results from three biological replicates and are the means ± SD. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (D) A Y1H assay shows that CsLOB1 interacts with the Cs9g12620 promoter PCs9g12620 and CsLOB1-binding site deletion mutants PCs9g12620-MLB1, PCs9g12620-MLB2, and PCs9g12620-MLB1/2. The upper diagram shows the putative CsLOB1-binding sites and the corresponding deletion mutants. The bar chart below represents a Y1H assay that shows the interaction of CsLOB1 with the promoter mutants. The analyses were same as those conducted in A. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (E) An EMSA showed that CsLOB1 binds to Cs9g12620-MLB1, PCs9g12620-MLB2, and PCs9g12620-MLB1/2. Purified GST-CsLOB1 was incubated with 25 ng of promoter DNA in gel shift binding buffer at 28°C for 30 min. The analyses were same as those in B. (F) Luciferase assays show the role of CsLOB1 on PCs9g12620-MLB1, PCs9g12620-MLB2, and PCs9g12620-MLB1/2. Promoter luciferase fusions were co-expressed with CsLOB1 in N. benthamiana. Co-expression with the empty binary vector pHB was used as the control. Luciferase activity was measured with a CCD imaging system at 2 days post-agroinfiltration. The image on the right shows the quantification of luciferase signal using a microplate luminescence reader. Values are the means ± SD (n = 3 biological replicates, **p ˂ 0.01 [Student’s t-test]). (G) qRT-PCR analysis of the transcript level of Cs9g12620 in CsLOB1-silenced plants. CsLOB1 was silenced in Citrus sinensis using the citrus tristeza virus (CTV)-based gene silencing vector CTV33. The effective infection by CTV was evaluated by the expression of P23 gene harbored in CTV33. The silencing efficiency of CsLOB1 was verified by comparison with the levels of expression in the WT and empty CTV33-infected control plants. The reduced expression of Cs9g12620 is shown by comparison with the level of expression in the empty CTV33-infected plants. The qRT-PCR was performed as described in C. ANOVA, analysis of variance; CCD, charge-coupled device; GUS, β-glucuronidase; PAGE, polyacrylamide gel electrophoresis; qRT-PCR, real-time quantitative reverse transcription PCR; SD, standard deviation; WT, wild type.
-
Figure 4—source data 1
Excel file containing LacZ, qRT, and RLU raw data in Figure 4A, C, D, F, G.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Original file for the yeast one-hybrid assay in Figure 4A (pGBKT7-CsLOB1+pG221-PCs9g12620, pGBKT7-CsLOB1+pG221, pGBKT7+pG221-PCs9g12620, and pGBKT7+pG221).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data2-v1.zip
-
Figure 4—source data 3
PDF containing Figure 4A and original scans of the yeast one-hybrid assay (pGBKT7-CsLOB1+pG221-PCs9g12620, pGBKT7-CsLOB1+pG221, pGBKT7+pG221-PCs9g12620, and pGBKT7+pG221) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data3-v1.pdf
-
Figure 4—source data 4
Original file for the electrophoretic mobility shift assay in Figure 4B (GST-LOB1+PCs9g12620).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data4-v1.zip
-
Figure 4—source data 5
PDF containing Figure 4B and original scans of the electrophoretic mobility shift assay (GST-LOB1+PCs9g12620) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data5-v1.pdf
-
Figure 4—source data 6
Original file for histochemical staining of the Cs9g12620 promoter-driven GUS activity in Figure 4C (PCs9g12620-GUS, PCs9g12620-GUS+35S:CsLOB1, PCs9g12620-GUS+EV, and uninoculatied).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data6-v1.zip
-
Figure 4—source data 7
PDF containing Figure 4C and original scans of the histochemical staining of the Cs9g12620 promoter-driven GUS activity assay (PCs9g12620-GUS, PCs9g12620-GUS+35S:CsLOB1, PCs9g12620-GUS+EV, and uninoculatied) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data7-v1.pdf
-
Figure 4—source data 8
Original file for the yeast one-hybrid assay in Figure 4D (pGBKT7-CsLOB1+PCs9g12620, pGBKT7-CsLOB1+PCs9g12620-MLB1, pGBKT7-CsLOB1+PCs9g12620-MLB2, and pGBKT7-CsLOB1+PCs9g12620-MLB1/2).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data8-v1.zip
-
Figure 4—source data 9
Original file for the yeast one-hybrid assay in Figure 4D (pGBKT7+PCs9g12620, pGBKT7+PCs9g12620-MLB1, pGBKT7+PCs9g12620-MLB2, pGBKT7+PCs9g12620-MLB1/2, pGBKT7-CsLOB1+pG221, and pGBKT7+pG221).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data9-v1.zip
-
Figure 4—source data 10
PDF containing Figure 4D and original scans of the yeast one-hybrid assay (pGBKT7-CsLOB1+PCs9g12620, pGBKT7-CsLOB1+PCs9g12620-MLB1, pGBKT7-CsLOB1+PCs9g12620-MLB2, pGBKT7-CsLOB1+PCs9g12620-MLB1/2, pGBKT7+PCs9g12620, pGBKT7+PCs9g12620-MLB1, pGBKT7+PCs9g12620-MLB2, pGBKT7+PCs9g12620-MLB1/2, pGBKT7-CsLOB1+pG221, and pGBKT7+pG221) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data10-v1.pdf
-
Figure 4—source data 11
Original file for the electrophoretic mobility shift assay in Figure 4E (GST-LOB1+PCs9g12620-MLB1 and GST-LOB1+PCs9g12620-MLB2).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data11-v1.zip
-
Figure 4—source data 12
Original file for the electrophoretic mobility shift assay in Figure 4E (GST-LOB1+PCs9g12620-MLB1/2).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data12-v1.zip
-
Figure 4—source data 13
PDF containing Figure 4E and original scans of the electrophoretic mobility shift assay (GST-LOB1+PCs9g12620-MLB1, GST-LOB1+PCs9g12620-MLB2, and GST-LOB1+PCs9g12620-MLB1/2) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data13-v1.pdf
-
Figure 4—source data 14
Original file for the luciferase assays in Figure 4F (PCs9g12620, PCs9g12620-MLB1, PCs9g12620-MLB2, and PCs9g12620-MLB1/2 co-expression with CsLOB1 and EV, respectively).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data14-v1.zip
-
Figure 4—source data 15
PDF containing Figure 4F and original scans of the luciferase assays (PCs9g12620, PCs9g12620-MLB1, PCs9g12620-MLB2, and PCs9g12620-MLB1/2 co-expression with CsLOB1 and EV, respectively) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig4-data15-v1.pdf
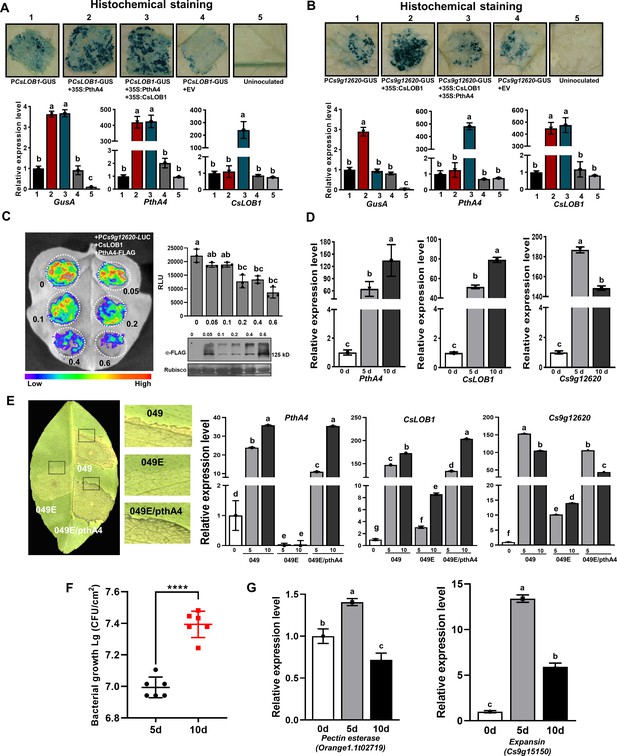
CsLOB1-induced Cs9g12620 promoter activity is suppressed by PthA4.
(A) PthA4 induced the activity of CsLOB1 promoter. PthA4 was transiently co-expressed with the CsLOB1 promoter GUS fusion in Nicotiana benthamiana. GUS activity was assayed at 2 days post-agroinfiltration. The upper images show the histochemical staining of CsLOB1 promoter (PCsLOB1)-driven GUS activity. The bar chart below shows the qRT-PCR analysis of the transcript levels of gusA, CsLOB1, and pthA4. The transcript level of each gene in the samples that expressed PCsLOB1-GUS was set as 1, and the levels of the other samples were calculated relative to that. Values are the mean results from three biological replicates and are the means ± SD. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (B) PhtA4 suppressed the CsLOB1-induced activity of the Cs9g12620 gene promoter. The suppression was evaluated by the co-expression of PthA4 with CsLOB1 and PCs9g12620-GUS. Histochemical staining and a qRT-PCR analysis were performed as described in A. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (C) PthA4 suppressed the CsLOB1-induced PCs9g12620 activity in a dose-dependent manner. CsLOB1 and PCs9g12620-LUC were co-expressed with PthA4-FALG in N. benthamiana. The agrobacteria that expressed PthA4-FLAG were arranged to a cell concentration series of OD600 values of 0.05, 0.1, 0.2, 0.4, and 0.6. The luciferase signal was quantified with a microplate luminescence reader. Values are the means ± SD (n = 3 biological replicates). Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). The image on the bottom right shows the expression of PthA4-FLAG by immunoblotting with anti-FLAG. (D) A qRT-PCR analysis of the levels of expression of pthA4, CsLOB1, and Cs9g12620 in Citrus sinensis leaves inoculated with Xcc 29-1. The cell suspension (108 CFU/ml) was infiltrated into the plant leaves. qRT-PCR was performed at 0, 5, and 10 days post-inoculation (dpi). The level of expression of each gene at 0 dpi was set as 1, and the level in other samples was calculated relative to those baseline values. Values are the mean results from three biological replicates and are the means ± SD. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (E) The level of expression of Cs9g12620 was dynamically related with that of PthA4 during Xcc infection. The disease symptoms caused by WT Xcc 049, transcriptional activator-like (TAL)-free mutant 049E, and Xcc 049E/pthA4 on C. sinensis leaves. The phenotype was recorded at 10 dpi. A qRT-PCR analysis was conducted to evaluate the levels of expression of pthA4, CsLOB1, and Cs9g12620. The experiment was performed as described in D. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). (F) Growth of wildtype Xcc 29-1 in C. sinensis plants. The Xcc 29-1 cells of 108 CFU/ml were infiltrated into citrus leaves. At 5 and 10 days post inoculation, bacteria were recovered from leaves and counted on Nutrient Agar (NA) plates. Error bars represent the standard deviation from three independent experiments (Student’s t-test, ****p < 0.0001). (G) qRT-PCR analysis of pectin esterase (orange1.1t02719) and expansin (cs9g15150) genes expression levels in C. sinensis leaves inoculated with Xcc 29-1. The experiment was performed as described in D. Columns labeled with different letters indicate significant difference among means (ANOVA, p < 0.01). ANOVA, analysis of variance; GUS, β-glucuronidase; qRT-PCR, real-time quantitative reverse transcription PCR; SD, standard deviation.
-
Figure 5—source data 1
Excel file containing qRT, RLU, and bacterial growth raw data in Figure 5A–G.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Original file for histochemical staining of the CsLOB1 promoter-driven GUS activity in Figure 5A (PCsLOB1-GUS, PCsLOB1-GUS+35S:PthA4, PCsLOB1-GUS+35S:PthA4+35S:CsLOB1, PCsLOB1-GUS+EV, and uninoculatied).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data2-v1.zip
-
Figure 5—source data 3
PDF containing Figure 5A and original scans of the histochemical staining of the CsLOB1 promoter-driven GUS activity assay (PCsLOB1-GUS, PCsLOB1-GUS+35S:PthA4, PCsLOB1-GUS+35S:PthA4+35S:CsLOB1, PCsLOB1-GUS+EV, and uninoculatied) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data3-v1.pdf
-
Figure 5—source data 4
Original file for histochemical staining of the Cs9g12620 promoter-driven GUS activity in Figure 5B (PCs9g12620-GUS, PCs9g12620-GUS+35S:CsLOB1, P Cs9g12620-GUS+35S:CsLOB1+35S:PthA4, PCs9g12620-GUS+EV, and uninoculatied).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data4-v1.zip
-
Figure 5—source data 5
PDF containing Figure 5B and original scans of the histochemical staining of the Cs9g12620 promoter-driven GUS activity assay (PCs9g12620-GUS, PCs9g12620-GUS+35S:CsLOB1, P Cs9g12620-GUS+35S:CsLOB1+35S:PthA4, PCs9g12620-GUS+EV, and uninoculatied) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data5-v1.pdf
-
Figure 5—source data 6
Original file for the luciferase assays in Figure 5C (PCs9g12620+CsLOB1+PthA4-FLAG).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data6-v1.zip
-
Figure 5—source data 7
PDF containing Figure 5C and original scans of the luciferase assays (PCs9g12620+CsLOB1+PthA4-FLAG) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data7-v1.pdf
-
Figure 5—source data 8
Original file for the western blot analysis in Figure 5C (anti-FLAG).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data8-v1.zip
-
Figure 5—source data 9
Original file for the western blot analysis in Figure 5C (Rubisco).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data9-v1.zip
-
Figure 5—source data 10
PDF containing Figure 5C and original scans of the western blot analysis (anti-FLAG and Rubisco) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data10-v1.pdf
-
Figure 5—source data 11
Original image for the disease symptoms analysis in Figure 5E (049, 049E, and 049E/pthA4).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data11-v1.zip
-
Figure 5—source data 12
PDF containing Figure 5E and original scans of the disease symptoms analysis (049, 049E, and 049E/pthA4) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig5-data12-v1.pdf
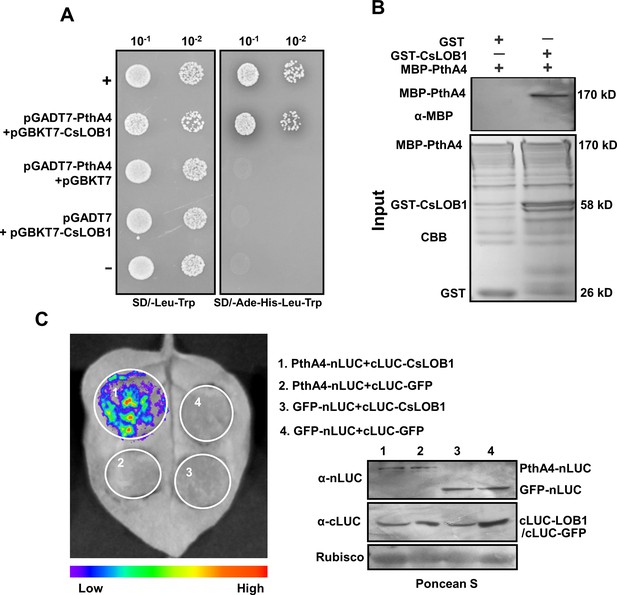
PhtA4 interacts with CsLOB1.
(A) A yeast two-hybrid assay shows that PhtA4 interacts with CsLOB1. pGADT7-PthA4 and pGBKT7-CsLOB1 were co-transformed in yeast cells and screened on synthetic dextrose media that lacked leucine and tryptophan (SD/-Leu-Trp). A single yeast colony was then selected for serial dilution and grown on SD/-Leu-Trp and SD/-Ade-His-Leu-Trp to examine the interaction. The yeast co-transformed with pGADT7-T and pGBKT7-53 served as a positive control. The yeast co-transformed with pGADT7-T and pGBKT7-lam served as a negative control. (B) GST pull-down assay of the interaction between PhtA4 and CsLOB1. The recombinant GST-CsLOB1 and MBP-PhtA4 proteins were used for GST pull-down assays. The GST protein served as a negative control. The pull-down of MBP-PhtA4 was verified by anti-MBP immunoblotting. The experiment was repeated three times with similar results. (C) A split luciferase assay for the interaction of PhtA4 and CsLOB1 in vivo. Nicotiana benthamiana leaves were co-infiltrated with agrobacteria that harbored 35S:PhtA4-nLUC and 35S:cLUC-CsLOB1. The image on the right shows the expression of respective proteins. Images of chemiluminescence were obtained by treatment with 0.5 μM luciferin 48 hr post-infiltration. Similar results were obtained in three biological repeats. GUS, β-glucuronidase.
-
Figure 6—source data 1
Original file for the yeast two-hybrid assay in Figure 6A (PthA4+CsLOB1, SD/-Leu-Trp).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data1-v1.zip
-
Figure 6—source data 2
Original file for the yeast two-hybrid assay in Figure 6A (PthA4+CsLOB1, SD/-Ade-His-Leu-Trp).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data2-v1.zip
-
Figure 6—source data 3
PDF containing Figure 6A and original scans of the yeast two-hybrid assay (PthA4+CsLOB1, SD/-Leu-Trp and SD/-Ade-His-Leu-Trp) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data3-v1.pdf
-
Figure 6—source data 4
Original file for the western blot analysis in Figure 6B (anti-MBP).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data4-v1.zip
-
Figure 6—source data 5
Original file for the Coomassie Brilliant Blue (CBB) analysis in Figure 6B.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data5-v1.zip
-
Figure 6—source data 6
PDF containing Figure 6B and original scans of the western blot analysis (anti-FLAG and CBB) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data6-v1.pdf
-
Figure 6—source data 7
Original file for the luciferase assays in Figure 6C (PthA4-nLUC+cLUC-CsLOB1, PthA4-nLUC+cLUC-GFP, GFP-nLUC+cLUC-CsLOB1, and GFP-nLUC+cLUC-GFP).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data7-v1.zip
-
Figure 6—source data 8
PDF containing Figure 6C and original scans of the luciferase assays (PthA4-nLUC+cLUC-CsLOB1, PthA4-nLUC+cLUC-GFP, GFP-nLUC+cLUC-CsLOB1, and GFP-nLUC+cLUC-GFP) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data8-v1.pdf
-
Figure 6—source data 9
Original file for the western blot analysis in Figure 6C (anti-nLUC and anti-cLUC).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data9-v1.zip
-
Figure 6—source data 10
Original file for the western blot analysis in Figure 6C (Rubisco).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data10-v1.zip
-
Figure 6—source data 11
PDF containing Figure 6C and original scans of the western blot analysis (anti-nLUC, anti-cLUC, and Rubisco) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig6-data11-v1.pdf
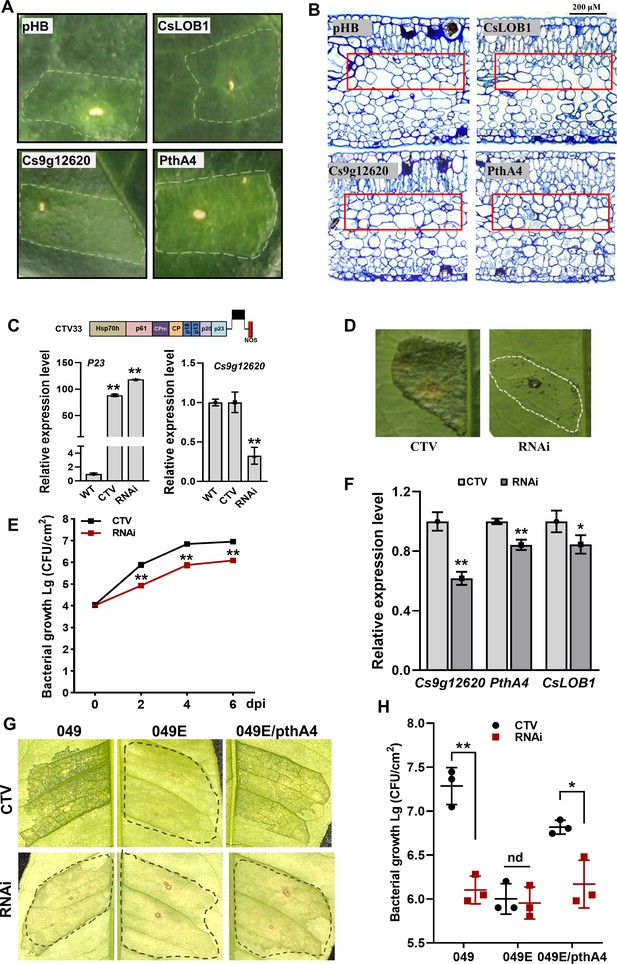
Cs9g12620 is involved in the formation of canker symptoms.
(A) The transient overexpression of Cs9g12620 results in hypertrophy in the Citrus sinensis leaves. The expression of pthA4 was used as the positive control. The cultured Agrobacterium cells were adjusted to an OD600 of 0.01 and infiltrated into young C. sinensis leaves. The phenotypes were recorded at 15 days post-agroinfiltration. The area of infiltration for each gene is indicated by a dotted line. (B) Transmission electron micrograph of C. sinensis leaf tissues. Transmission electron micrographs of the cross sections of citrus leaf tissue. The leaves were sampled at 15 days post-agroinfiltration. The changes in tissue structure induced by Cs9g12620 and pthA4 are indicated by a box. The bars represent 200 μM. (C) A real-time quantitative reverse transcription PCR (qRT-PCR) analysis of the silencing of Cs9g12620 in C. sinensis. Cs9g12620 was silenced using the citrus tristeza virus (CTV)-based gene silencing vector CTV33. The effective infection by CTV was evaluated by measuring the level of expression of the P23 gene harbored in CTV33. The silencing efficiency of Cs9g12620 was verified by comparison with the levels of expression in the WT and empty CTV33-infected control plants. Values are the mean results from three biological replicates (Student’s t-test, **p < 0.01). The experiment was repeated three times. (D) The canker symptoms on Cs9g12620-silenced citrus plants were caused by the WT Xcc 29-1. The Xcc 29-1 cells of 107 CFU/ml were infiltrated into citrus leaves to analyze the development of canker symptoms. The canker symptoms were recorded at 6 days post-inoculation (dpi). The areas of infiltration are indicated by dotted lines. The inoculation of the plants infected by the empty vector CTV33 was used as the negative control. (E) Growth of Xcc 29-1 in Cs9g12620-silenced citrus plants. A volume of 107 CFU/ml of Xcc 29–1 cells were infiltrated into citrus leaves. At 2, 4, and 6 dpi, bacteria were recovered from leaves and counted on Nutrient Agar (NA) plates. Error bars represent the standard deviation from three independent experiments (Student’s t-test, *p < 0.05, **p < 0.01). (F) A qRT-PCR analysis of Cs9g12620, CsLOB1, and pthA4 in Cs9g12620-silenced citrus plants inoculated with Xcc 29-1. The experiment was performed as described in C. (G) The canker symptoms on Cs9g12620-silenced citrus plants were caused by the WT Xcc 049, transcriptional activator-like (TAL)-free mutant 049E, and 049E/PthA4. The experiment was performed as described in D. (H) Growth of WT Xcc 049, 049E, and 049E/PthA4 in Cs9g12620-silenced citrus plants. At 6 dpi, bacteria were recovered from leaves and counted on nutrient-rich (NA) plates. The experiment was performed as described in E. dpi, days post-inoculation; WT, wild type.
-
Figure 7—source data 1
Excel file containing qRT and bacterial growth raw data in Figure 7C, E, F, H.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data1-v1.xlsx
-
Figure 7—source data 2
Original image of transient overexpression (pHB, CsLOB1, Cs9g12620, and PthA4) in the C. sinensis leaves in Figure 7A.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data2-v1.zip
-
Figure 7—source data 3
PDF containing Figure 7A and original scans of the transient overexpression analysis (pHB, CsLOB1, Cs9g12620, and PthA4) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data3-v1.pdf
-
Figure 7—source data 4
Original image of transmission electron micrograph of C. sinensis leaf tissues transient overexpression pHB in Figure 7B.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data4-v1.zip
-
Figure 7—source data 5
Original image of transmission electron micrograph of C. sinensis leaf tissues transient overexpression CsLOB1 in Figure 7B.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data5-v1.zip
-
Figure 7—source data 6
Original image of transmission electron micrograph of C. sinensis leaf tissues transient overexpression Cs9g12620 in Figure 7B.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data6-v1.zip
-
Figure 7—source data 7
Original image of transmission electron micrograph of C. sinensis leaf tissues transient overexpression PthA4 in Figure 7B.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data7-v1.zip
-
Figure 7—source data 8
Original image for the disease symptoms analysis in Figure 7D (citrus tristeza virus, CTV).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data8-v1.zip
-
Figure 7—source data 9
Original image for the disease symptoms analysis in Figure 7D (RNAi).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data9-v1.zip
-
Figure 7—source data 10
Original image for the disease symptoms analysis in Figure 7G (citrus tristeza virus, CTV).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data10-v1.zip
-
Figure 7—source data 11
Original image for the disease symptoms analysis in Figure 7G (RNAi).
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data11-v1.zip
-
Figure 7—source data 12
PDF containing Figure 7G and original scans of the disease symptoms analysis (citrus tristeza virus [CTV] and RNAi) with highlighted bands.
- https://cdn.elifesciences.org/articles/91684/elife-91684-fig7-data12-v1.pdf
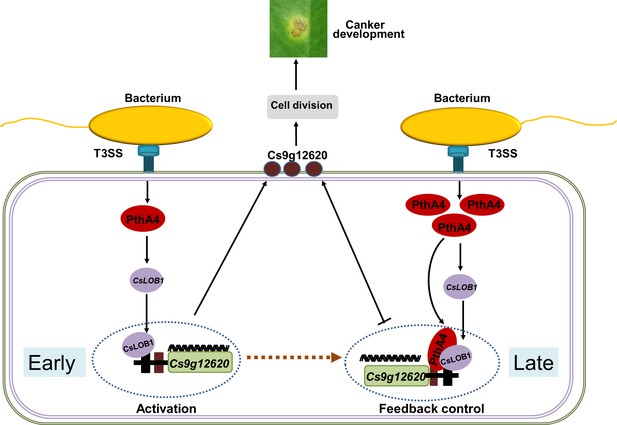
Proposed model of the expression of Cs9g12620 directed by PthA4-mediated induction of CsLOB1.
The regulation of putative carbohydrate-binding protein gene Cs9g12620 was divided into two steps. At the early infection stage, the presence of PthA4 is responsible for the induction of CsLOB1, which is responsible for the induction of Cs9g12620. At the late infection stage, the over-redundancy of PthA4 exerts a feedback suppression effect on the activation of Cs9g12620 by interacting with CsLOB1.
Additional files
-
Supplementary file 1
Supplementary file with additional data.
(a) List of bacterial strains and plasmids used in this study. (b) List of primers used in this study.
- https://cdn.elifesciences.org/articles/91684/elife-91684-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91684/elife-91684-mdarchecklist1-v1.docx





