Netrin signaling mediates survival of dormant epithelial ovarian cancer cells
Figures
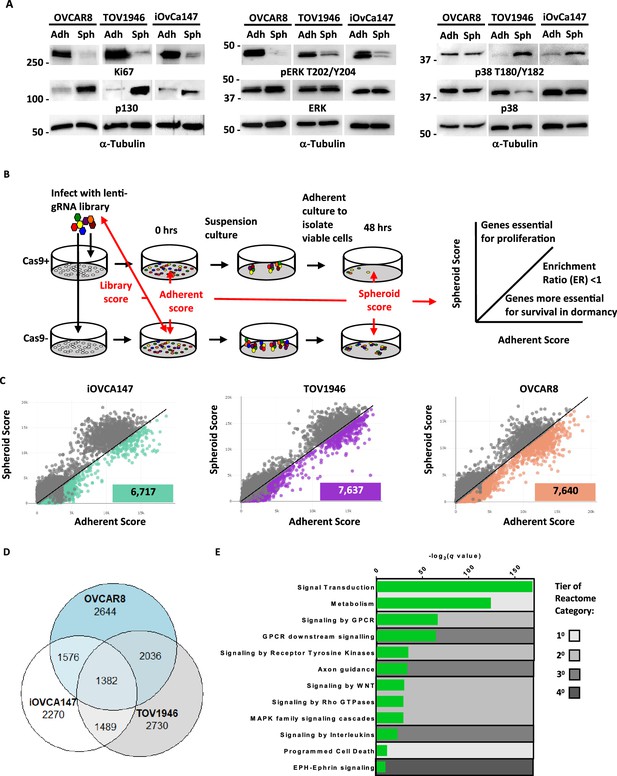
GO-CRISPR screens implicate axon guidance pathways as supporting HGSOC spheroid cell viability.
(A) iOvCa147, TOV1946, or OVCAR8 cells were cultured under adherent conditions (Adh) or in suspension to induce spheroid formation (Sph). Lysates were prepared and analyzed by western blotting for the proliferation marker Ki67 and the quiescence marker p130, and phosphorylated and total levels of ERK and p38. Tubulin was blotted as a loading control. (B) Flow chart of GO-CRISPR screening used for each of iOvCa147, TOV1946, or OVCAR8 control cells and derivatives of each that express Cas9. Cas9-positive cells (top row) and Cas9-negative cells (bottom row) were transduced with the GeCKO v2 pooled sgRNA library. After antibiotic selection, cells were expanded under adherent culture conditions (0 hr) before being transferred to suspension culture conditions to induce spheroid formation and select for cell survival. After 48 hr, spheroids were transferred to standard plasticware to isolate viable cells. Red arrows indicate the relevant comparisons of sgRNA sequence abundance that were made to analyze screen outcomes. Genes with relatively greater effect on viability in suspension were selected by comparing their scores between adherent and suspension conditions and considering genes with an enrichment ratio (ER) of <1. (C) Scatter plots representing the spheroid score on y-axis and adherent score on the x-axis calculated by TRACS for each gene in each cell line (iOvCa147, TOV1946, OVCAR8). Colored data points represent genes with ER <1 and padj <0.05 (paired t-test). (D) Venn diagram illustrating overlap of genes identified as supporting cell viability in suspension culture from iOvCa147, TOV1946, and OVCAR8 cells. (E) Graph depicting enriched pathways from ConsensusPathDB using the 1382 commonly identified genes from D. Categories are ranked by q-value. Tiers of Reactome categories are indicated by shading.
-
Figure 1—source data 1
Original files for western blot analysis in Figure 1A (Ki67, p130, pERK T202/Y204, ERK, p38 T180/Y182, p38, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig1-data1-v1.zip
-
Figure 1—source data 2
PDF containing annotation of original western blots in Figure 1A (Ki67, p130, pERK T202/Y204, ERK, p38 T180/Y182, p38, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig1-data2-v1.pdf
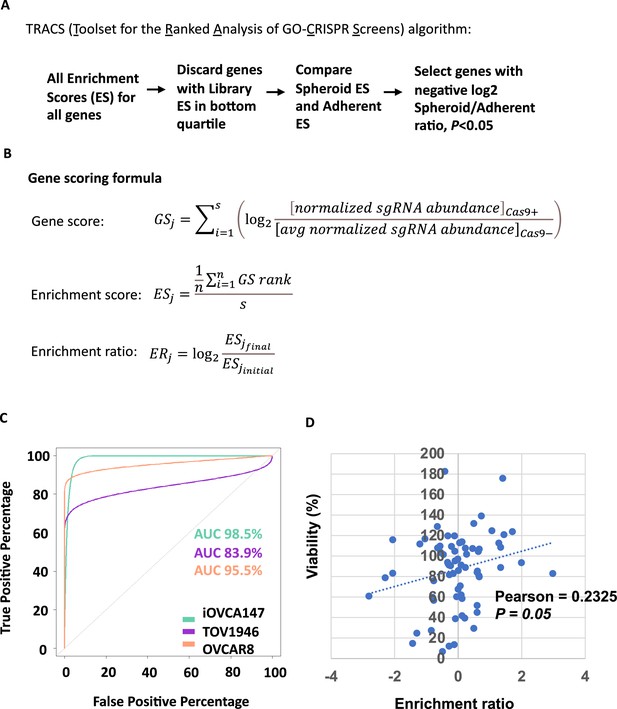
CRISPR screen analysis details and internal controls.
(A) Flowchart of data analysis used to identify genes that are most relevant to ovarian cancer cell survival in suspension. (B) Mathematical formulas to define gene scores, enrichment scores (n=guides per gene), and enrichment ratios used by TRACS. (C) Reader-operator curves were generated to assess TRACS categorization of 1000 non-targeting control sgRNAs in the GeCKOv2 library for each of the three cell lines screened. (D) Spheroid cell viability levels from low throughput siRNA gene knock downs for 35 genes in iOvCa147 and OVCAR8 cells was plotted against the enrichment ratio for the same genes in the same two cell lines from this screen. These independent approaches to measure viability were compared using linear regression.
-
Figure 1—figure supplement 1—source data 1
Numerical data used in the graph in Figure 1—figure supplement 1D.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig1-figsupp1-data1-v1.xlsx
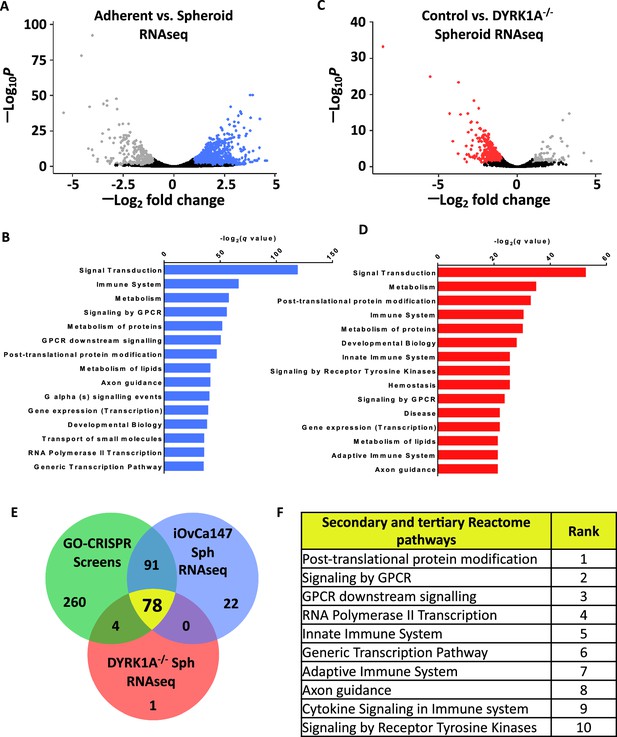
Axon guidance pathway components are upregulated in iOvCa147 spheroid cells in a DYRK1A dependent manner.
(A) RNA was isolated from iOvCa147 cells following culture under adherent conditions or in suspension conditions to induce spheroid formation for 6 hr. Triplicate independent cultures were processed for RNA-seq. A volcano plot shows differentially expressed genes in spheroid cells compared to adherent. 1937 genes were found to be downregulated in iOvCa147 spheroid cells (log2 fold change <1, padj <0.05 (Wald test), FDR 10%, highlighted in grey) and 1,834 genes were upregulated (log2 fold change >1, padj <0.05, FDR 10%, highlighted in blue). (B) Top 15 most significantly enriched pathways (padj <0.05) whose genes were upregulated in suspension culture compared to adherent in RNA-seq analysis. (C) A volcano plot showing differentially expressed genes in DYRK1A-/- spheroid cells compared to iOvCa147 spheroid cells. A ttoal of 744 genes were found to be downregulated in DYRK1A-/- spheroid cells (log2 fold change <1, padj <0.05 (Wald test), FDR 10%, highlighted in red) and 96 genes were upregulated (log2 fold change >1, padj <0.05 (Wald test), FDR 10%, highlighted in grey). (D) Top 15 most significantly enriched pathways that were represented by downregulated genes in DYRK1A-/- suspension culture compared to control cells in suspension. (E) Venn diagram depicting overlapping enriched pathways identified in GO-CRISPR screens in green; enriched pathways identified in upregulated genes in parental iOvCa147 spheroid cells in blue; and enriched pathways identified in downregulated genes in DYRK1A-/- spheroid cells in red. 78 pathways were commonly enriched in all three datasets (shown in yellow). (F) Top 10 most significantly enriched pathways among the 78 identified in C.
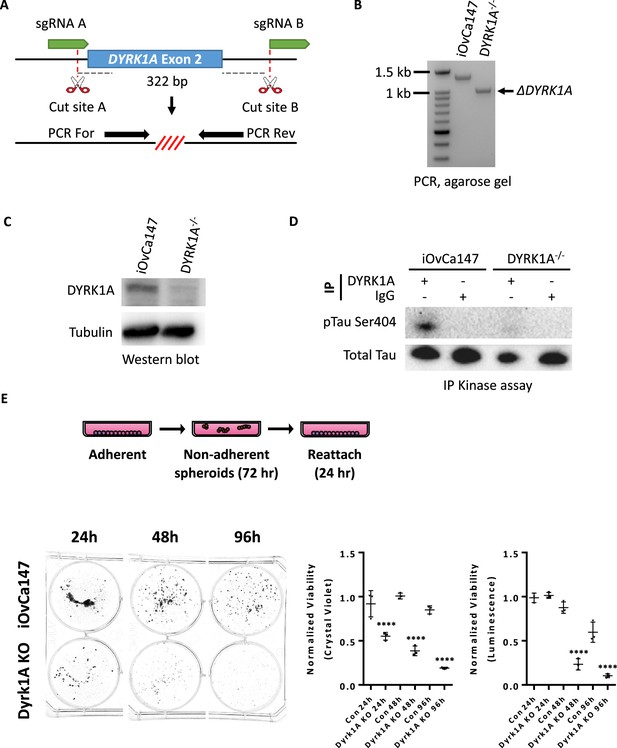
Generation of iOvCa147 cells deficient for DYRK1A and loss of viability in suspension.
(A) Strategy to generate DYRK1A-/- cells using a pair of sgRNAs that flank exon 2. PCR For and PCR Rev primers flank exon 2 and are used to detect deletion events. (B) Agarose gel showing PCR products for iOVCA147 cells and putative DYRK1A-/- cells. The full length amplicon containing exon 2 was detected in parental iOvCa147 cells (1348 bp). A smaller amplicon (1026 bp) was detected in DYRK1A-/- cells, indicting successful excision of the 322 bp region encompassing exon 2. (C) Western blot comparing DYRK1A expression in iOVCA147 cells and DYRK1A-/- cells. (D) IP kinase assay to evaluate DYRK1A activity in DYRK1A-/- cells. Anti-DYRK1A antibodies or IgG was used to immunoprecipitate from iOVCA147 and DYRK1A-/- cells. Precipitates were incubated with ATP and Tau protein. Samples were resolved by SDS-PAGE and probed with pSer404-Tau antibody. (E) Parental iOvCa147 or DYRK1A-/- cells were incubated in suspension conditions for 24 hr, 72 hr, or 4 days to induce spheroid formation, and then re-plated in adherent conditions for 24 hr to allow reattachment. Reattached spheroid cells were stained with crystal violet and absorbance was quantified. Alternatively, cells in suspension were isolated and utilized for Cell TitreGLO cell viability measurements. DYRK1A-/- spheroid cells had impaired survival compared to parental iOvCa147 spheroid cells at each time point and reattached and crystal violet stained cell viability matched Cell TitreGLO measurements of viability (one-way anova, ****p<0.0001).
-
Figure 2—figure supplement 1—source data 1
Original files for western blot analysis in Figure 2—figure supplement 1C and D – (DYRK1A, pTAU S404, TAU, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
PDF containing annotation of original western blots in Figure 2—figure supplement 1C and D – (DYRK1A, pTAU S404, TAU, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig2-figsupp1-data2-v1.pdf
-
Figure 2—figure supplement 1—source data 3
Numerical data used in the graph in Figure 2—figure supplement 1E.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig2-figsupp1-data3-v1.xlsx
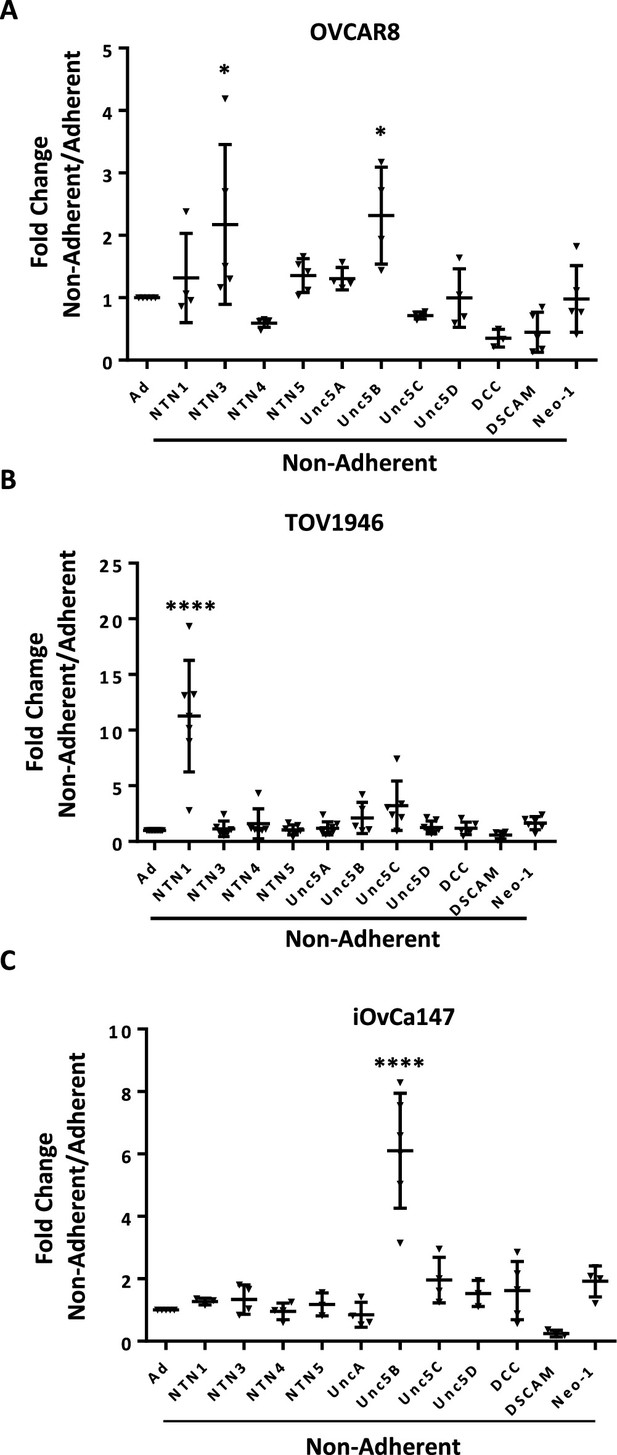
Expression of Netrin ligands and their dependence receptors is increased in suspension culture.
(A–C) RT-qPCR was performed to quantitate mRNA expression levels of Netrin ligands and receptors in three different HGSOC cell lines. Relative expression of the indicated transcripts is shown for suspension culture conditions compared with adherent. All experiments were performed in at least triplicate biological replicates. Means were compared with the same gene in adherent culture using a one way anova (*p<0.05, ****p<0.0001).
-
Figure 3—source data 1
Numerical data used in the graph in Figure 3.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig3-data1-v1.xlsx
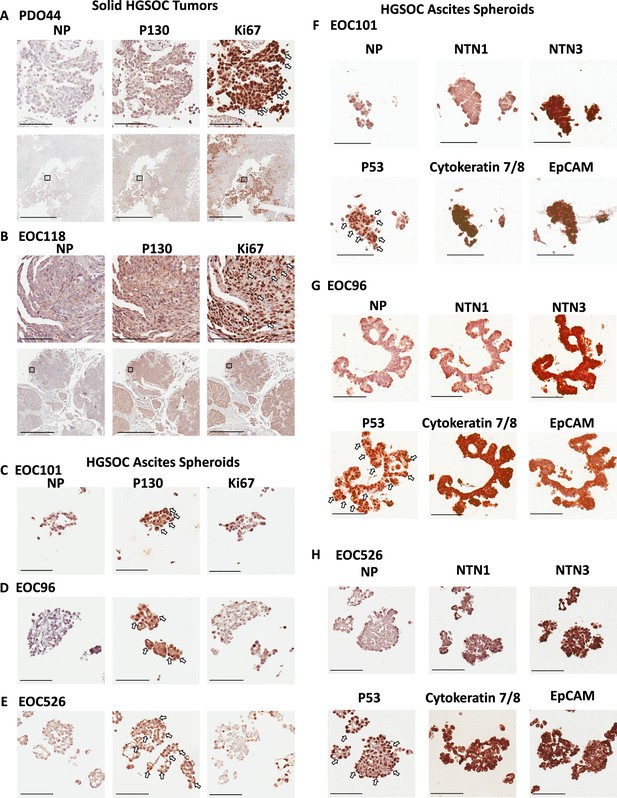
Netrins are expressed in dormant patient derived spheroids.
Spheroids were isolated from ascites of HGSOC patients and processed for immunohistochemical staining. (A–B) Serial sections from solid HGSOC tumors were stained with antibodies to p130, Ki67, or a no primary antibody control (NP). Scale bar = 100 μm for upper panels and 2 mm for lower panels. (C–E) Serial spheroid sections from the indicated patients were stained with Ki67 and p130. Sections with dense positive nuclear staining are indicated with white arrows. Scale bar = 100 μm. (F–H). Immunohistochemical staining was performed for the indicated proteins on serial sections of ascites derived spheroids. Omission of primary antibody was used a control for background staining for each patient sample (NP). Scale bar = 100 μm.
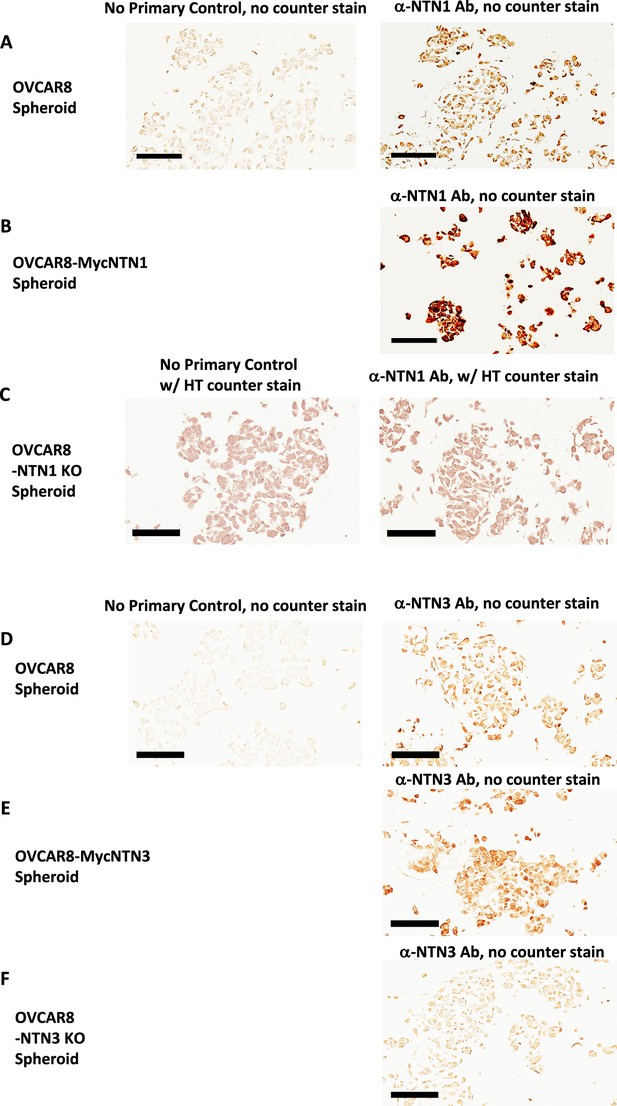
Specificity of Netrin-1 and Netrin-3 IHC staining of OVCAR8 spheroids.
(A) OVCAR8 cells were allowed to form spheroids for 6 hr before fixation in formalin and embedding in paraffin for sectioning. Serial sections were stained with Netrin-1 Abs or a no primary antibody control. (B) OVCAR8 cells overexpressing Netrin-1 were used to prepare spheroids as in A and stained in parallel to samples in A. (C) OVCAR8 deleted for Netrin-1 expression were used to prepare spheroids as in A and serial sections were stained for Netrin-1 or with a no primary antibody control and counter stained. (D) OVCAR8 cells were allowed to form spheroids for 6 hr before fixation in formalin and embedding in paraffin for sectioning. Serial sections were stained with Netrin-3 Abs or a no primary antibody control. (E) OVCAR8 cells overexpressing Netrin-3 were used to prepare spheroids as in D and stained in parallel to samples in D. (F) OVCAR8 deleted for Netrin-3 expression were used to prepare spheroids as in D and serial sections were stained for Netrin-3 or with a no primary antibody control. All scale bars = 100 μm.
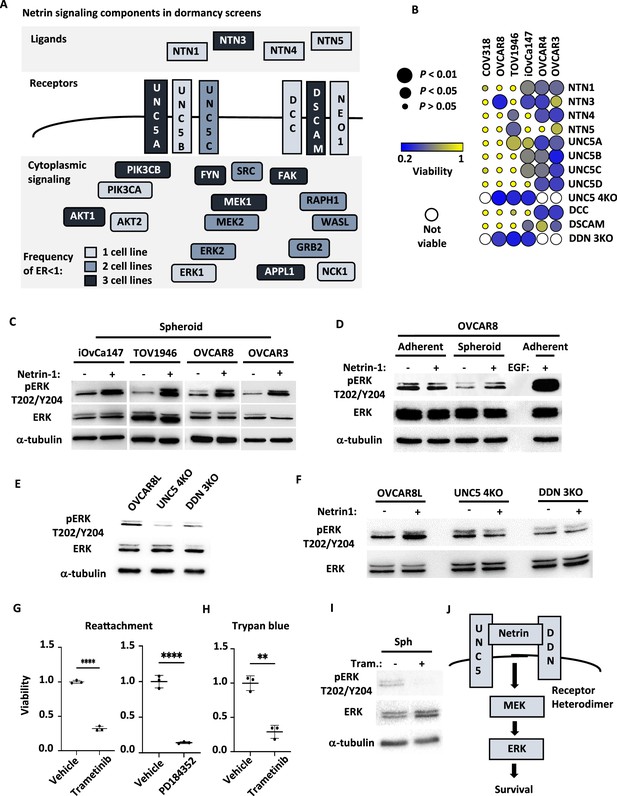
Netrin ligands and their receptors are required for spheroid cell survival.
(A) Illustration of netrin ligands, receptors, and other intracellular signaling molecules that are included in the Axon Guidance pathway category. The frequency of their identification in CRISPR screens is illustrated by shading and indicates how many cell lines had an enrichment ratio <1 for a given component. (B) The indicated ovarian cancer cell lines were infected with lentiviruses expressing sgRNAs directed against the indicated Netrin signaling genes. Cells were transferred to suspension culture conditions to induce spheroid formation for 72 hr and then returned to adherent conditions for 24 hr to facilitate reattachment. Re-attached cells were stained with Crystal Violet and retained dye was extracted and quantitated to measure relative survival. Each cell-gene combination was assayed in at least three biological replicates, averaged, and viability is displayed as a bubble plot. Mean survival for a given cell-gene combination was compared with GFP control gRNA transduced cells using one way anova and significance levels are illustrated by bubble size. Inviable cell-gene combinations are depicted as empty spaces. (C) Cultures of the indicated cell lines were cultured in suspension for five days and stimulated with 0.5 μg/mL Netrin-1. Netrin-1 signaling was analyzed by SDS-PAGE and western blotting for phospho-ERK, ERK, and tubulin. (D) Quiescent adherent OVCAR8 cells were stimulated with 0.5 μg/mL Netrin-1 or 0.5 μg/mL EGF, and compared with OVCAR8 cells in suspension stimulated as in C. Extracts were prepared and blotted for phospho-ERK, ERK, and tubulin. (E) Suspension cultures of OVCAR8, or knock out derivatives, were harvested and analyzed for relative phosphorylation levels of ERK by western blotting. Total ERK and tubulin blotting serve as expression and loading controls. (F) Netrin-1 signaling in OVCAR8, UNC5 4KO and DDN 3KO derivatives was tested by transferring cells to suspension and stimulating with Netrin-1 as before. Western blotting for phospho-ERK, ERK were also as before. (G) OVCAR8 cells were seeded in suspension culture and treated with the MEK inhibitors PD184352, Trametinib or DMSO vehicle for 72 hr. Mean viability was determined by re-attachment and compared by one way anova (****p<0.0001). (H) OVCAR8 cells were cultured in suspension and treated with Trametinib or DMSO vehicle as in G. Viability was determined by trypan blue dye exclusion and compared by one way anova (**p<0.01). (I) Extracts were prepared from Trametinib and control treated spheroid cells and blotted for phospho-ERK, ERK, and tubulin. (J) Model summarizing the roles of Netrin ligands, receptors, and downstream targets MEK and ERK in dormant survival signaling.
-
Figure 5—source data 1
Numerical data used for bubble plot in Figure 5B.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Original files for western blot analysis in Figure 5C (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data2-v1.zip
-
Figure 5—source data 3
PDFs containing annotation of original western blots in Figure 5C (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data3-v1.pdf
-
Figure 5—source data 4
Original files for western blot analysis in Figure 5D (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data4-v1.zip
-
Figure 5—source data 5
PDFs containing annotation of original western blots in Figure 5D (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data5-v1.pdf
-
Figure 5—source data 6
Original files for western blot analysis in Figure 5E (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data6-v1.zip
-
Figure 5—source data 7
PDFs containing annotation of original western blots in Figure 5E (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data7-v1.pdf
-
Figure 5—source data 8
Original files for western blot analysis in Figure 5F (pERK T202/Y204, ERK).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data8-v1.zip
-
Figure 5—source data 9
PDFs containing annotation of original western blots in Figure 5F (pERK T202/Y204, ERK).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data9-v1.pdf
-
Figure 5—source data 10
Numerical data used for graphs in Figure 5G and H.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data10-v1.xlsx
-
Figure 5—source data 11
Original files for western blot analysis in Figure 5E (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data11-v1.zip
-
Figure 5—source data 12
PDFs containing annotation of original western blots in Figure 5E (pERK T202/Y204, ERK, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-data12-v1.pdf
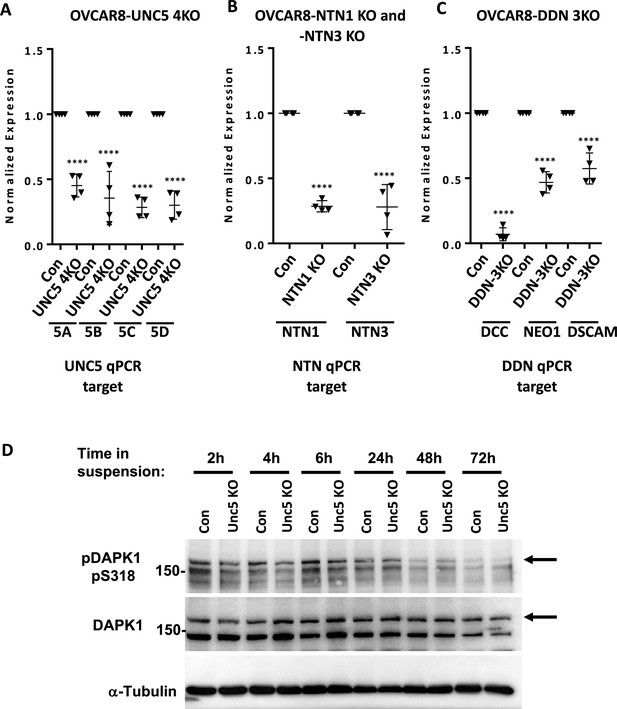
Evaluation of target gene transcript levels in multigene knock out cells.
(A) RT-qPCR was performed on RNA isolated from control sgRNA expressing OVCAR8 cells and UNC5 4KO to assess expression of UNC5 family members. Graphs depict relative expression levels for each indicted gene in control and knock out cell lines. Mean expression levels of targeted cells were compared to background measurements using one way anova (****p<0.0001). (B) RT-qPCR was used to measure mRNA levels of NTN1 and NTN3 transcripts in their respective CRISPR targeted, and OVCAR8 controls. Mean values were compared by one way anova (****p<0.0001). (C) Levels of mRNA corresponding to DCC, DSCAM, and NEO1 were determined by RT-qPCR in DDN 3KO cells and control OVCAR8. Mean values were compared by one way anova (****p<0.0001). (D) The indicated genotypes of cells were subjected to suspension culture and protein extracts were prepared at the indicated times. Proteins were resolved by SDS-PAGE, blotted, and probed with the indicated antibodies.
-
Figure 5—figure supplement 1—source data 1
Numerical data used for graphs in Figure 5—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-figsupp1-data1-v1.xlsx
-
Figure 5—figure supplement 1—source data 2
Original files for western blot analysis in Figure 5—figure supplement 1D (pDAPK1 S318, DAPK1, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-figsupp1-data2-v1.zip
-
Figure 5—figure supplement 1—source data 3
PDF containing annotation of original western blots in Figure 5—figure supplement 1D (pDAPK1 S318, DAPK1, Tubulin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig5-figsupp1-data3-v1.pdf
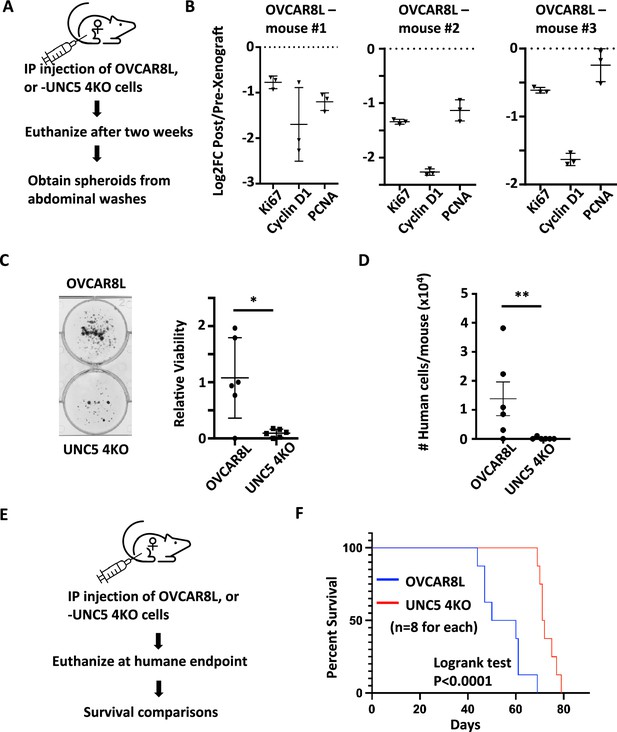
Loss of Netrin signaling reduces spheroids and prolongs survival.
(A) Control OVCAR8L (negative control CRISPR targeted cells) and UNC5 4KO cells were injected into the peritoneal space of female NOD/SCID mice. After 2 weeks, animals were euthanized and engrafted cells were collected with abdominal washes. (B) RT-qPCR was used to compare gene expression of the indicated cell cycle markers in RNA extracted from proliferating cells in culture before engraftment and compared with RNA obtained from spheroids in xenografted mice. Technical replicates of RNA derived from three different mice is shown. (C) Spheroids obtained from abdominal washes from each mouse were plated in adherent conditions to compare their abundance between control and UNC5 4KO genotypes. One example of each genotype of cell is shown. Crystal violet staining and dye extraction were used to quantitate biomass and averages were compared by one-way anova (n=6, *p<0.05). (D) DNA was extracted from cells collected in abdominal washes and human Alu repeats were detected by qPCR to quantitate and compare human cancer cells. Means were compared by one-way anova (n=6, **p<0.01). (E) Control OVCAR8 and UNC5 4KO cells were xenografted as above and mice were monitored until humane endpoint. (F) Kaplan-Meier analysis of survival for mice engrafted with the indicated genotypes of cells. Survival was compared by logrank test.
-
Figure 6—source data 1
Numerical data used for graphs in Figure 6B.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Numerical data used for graphs in Figure 6C.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig6-data2-v1.xlsx
-
Figure 6—source data 3
Numerical data used for graphs in Figure 6D.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig6-data3-v1.xlsx
-
Figure 6—source data 4
Numerical data used for graphs in Figure 6F.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig6-data4-v1.xlsx
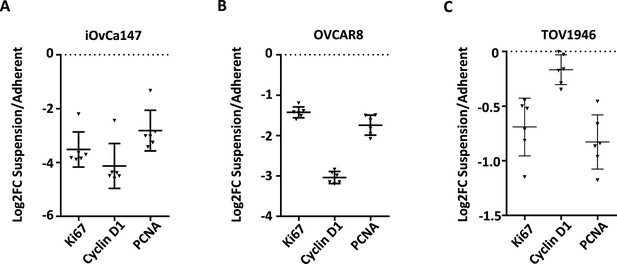
Evaluation of dormancy arrest in cell culture by RT-qPCR.
RNA was extracted from asynchronously proliferating cells and suspension cultures 72 hr post transfer from adherent. RT-qPCR was used to compare gene expression of the indicated cell cycle markers in iOvCa147, OVCAR8, TOV1946.
-
Figure 6—figure supplement 1—source data 1
Numerical data used for graphs in Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig6-figsupp1-data1-v1.xlsx
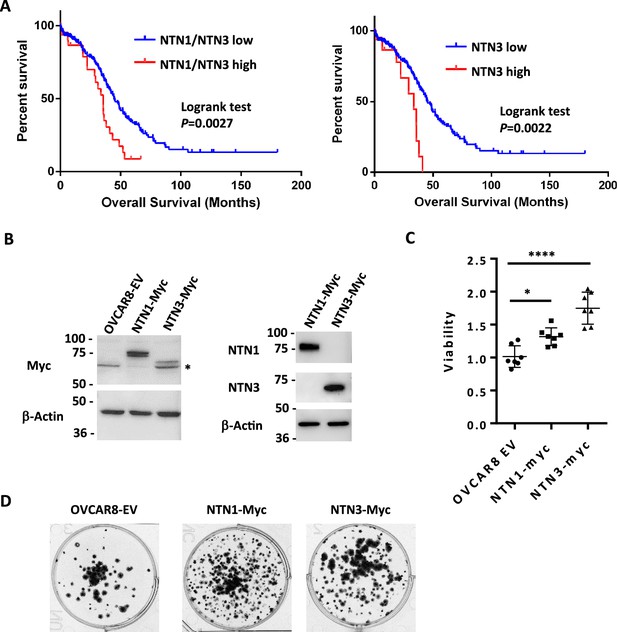
Netrin ligand overexpression is associated with poor clinical outcome in HGSOC.
(A) TCGA RNA-seq data for HGSOC patients (TCGA PanCancer Atlas study) was used to identify high Netrin-1 or –3 and low Netrin-1 or –3 expressing patients (high expressing are above z-score 1.2). Overall survival was used to construct Kaplan-Meier plots and survival was compared using a logrank test. (B) OVCAR8 cells were stably transduced with lentiviral constructs to overexpress epitope tagged Netrin-1 or –3. Western blotting for Netrins, Myc-tags, and Actin were used to determine relative expression levels of both Netrins in these cell populations and vector controls. (C) Control and Netrin overexpressing cells were transferred to suspension culture conditions to form spheroids, and replated to assay for viability. Mean viability and standard deviation is shown for each. One-way anova was used to compare survival (* p<0.05, *** p<0.001). (D) Reattached spheroids were fixed and stained with Crystal Violet to examine size and abundance in control and Netrin-1 or –3 overexpression.
-
Figure 7—source data 1
Original files for western blot analysis in Figure 7B (Myc, Netrin-1, Netrin-3, Actin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig7-data1-v1.zip
-
Figure 7—source data 2
PDFs containing annotation of original western blot analysis in Figure 7B (Myc, Netrin-1, Netrin-3, Actin).
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig7-data2-v1.pdf
-
Figure 7—source data 3
Numerical data used for graphs in Figure 7C.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig7-data3-v1.xlsx
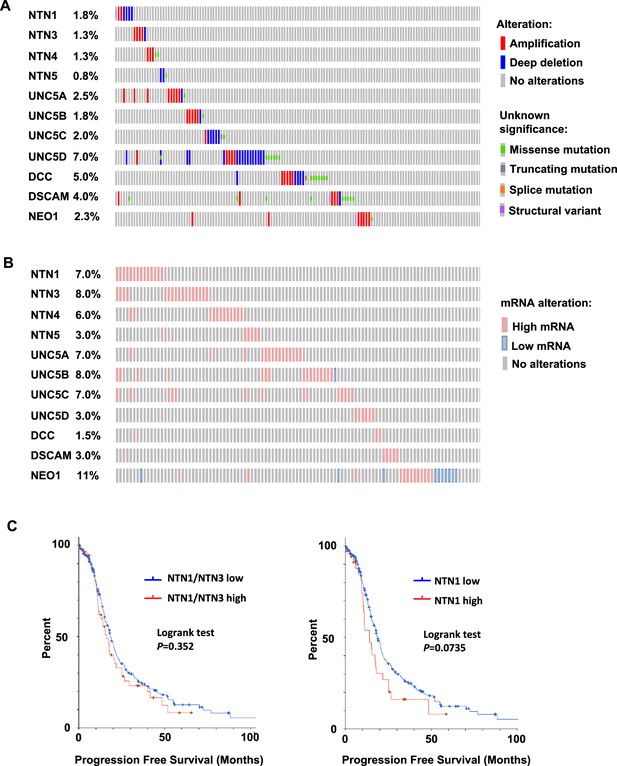
Frequency of Netrin ligand and receptor deletions, mutations or expression changes in high grade serous ovarian cancer.
(A) Oncoprint of genetic alterations in 10 netrin signaling related genes in 398 patients. Alteration types are defined on the right. Only the first 154 patients are shown. (B) Oncoprint illustrating gene expression outliers among 10 netrin signaling related genes in 201 patients (first 122 patients shown). Genomic data used is from Ovarian Serous Cystadenocarcinoma (TCGA, PanCancer Atlas) (201 samples with RNA seqV2, z-score cut off of 1.5). (C) TCGA RNA-seq data for HGSOC patients (TCGA PanCancer Atlas study) was used to identify high Netrin-1 or –3 and low Netrin-1 or –3 expressing patients (high expressing are above z-score 1.2). Progression free survival was used to construct Kaplan-Meier plots and survival was compared using a logrank test.
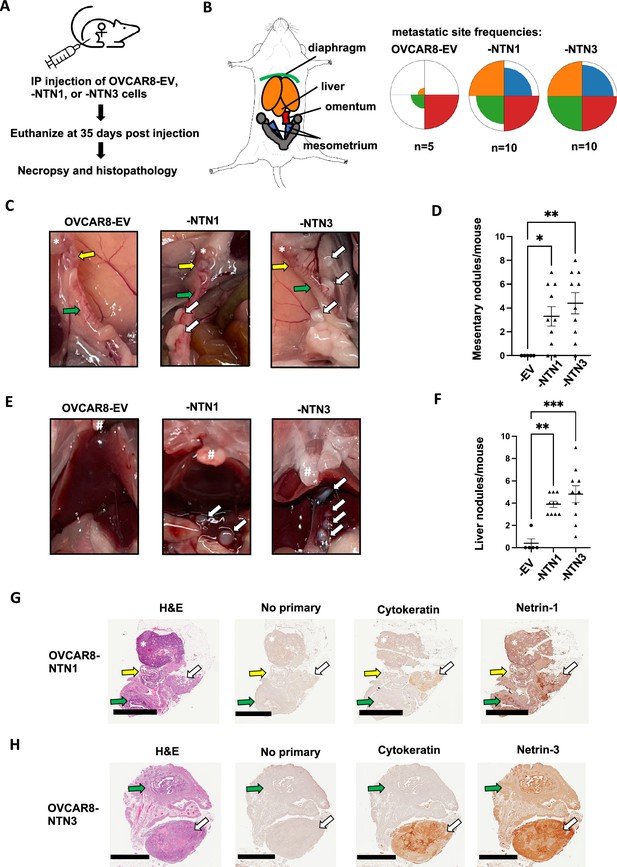
Netrin overexpression causes increased dissemination of tumor nodules.
(A) Netrin overexpressing and control OVCAR8 cells were injected into the intraperitoneal space of female NOD/SCID mice. Mice were euthanized following 35 days and analyzed for disease burden by necropsy and histopathology. (B) The spread of cancer to the diaphragm, liver, omentum, and mesometrium was determined from necropsies and colors used in the anatomical schematic correspond with petal plots for each genotype of cells. Petal plot radius illustrates frequency of mice bearing disease spread to a particular location. The radius of color fill is proportional to the total number of animals with tumor nodules found in that location. (C) Photographs of necropsy findings in the mesometrium. Locations of ovaries (*), oviduct (yellow arrow), and uterine horn (green arrow) are indicated in each case. Tumor nodules are indicated by white arrows. (D) The number of mesometrium associated tumor nodules was determined for each mouse. Mean values are indicated and differences between genotype were determined by one way anova (* p<0.05, ** p<0.01). (E) Photographs of necropsy findings in the liver. Location of sternum is indicated (#) in each image. Tumor nodules are indicated by white arrows. (F) The number of liver associated tumor nodules was determined for each mouse. Mean values are indicated and differences between genotype were determined by one way anova (** p<0.01, *** p<0.001). (G and H) Histology of mesometrial tumor nodules are shown. Serial sections were stained with H&E, or with the indicated antibodies for immunohistochemistry. Ovaries are indicated (*), as are the oviduct (yellow arrow), the uterus (green arrow), and the tumor nodule (white arrow). Scale bar = 2 mm.
-
Figure 8—source data 1
Numerical data used for graphs in Figure 8.
- https://cdn.elifesciences.org/articles/91766/elife-91766-fig8-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | NTN1 | GenBank | NCBI: 9423 Gene Cards: GC17P100538 | |
| Gene (Homo sapiens) | NTN3 | GenBank | NCBI: 4917 Gene Cards: GC17P100538 | |
| Gene (Homo sapiens) | NTN4 | GenBank | NCBI: 59277 Gene Cards: GC12M095657 | |
| Gene (Homo sapiens) | NTN5 | GenBank | NCBI: 126147 Gene Cards: GC19M048661 | |
| Gene (Homo sapiens) | Unc5A | GenBank | NCBI: 90249 Gene Cards: GC05P185698 | |
| Gene (Homo sapiens) | Unc5B | GenBank | NCBI: 219699 Gene Cards: GC10P071212 | |
| Gene (Homo sapiens) | Unc5C | GenBank | NCBI: 8633 Gene Cards: GC04M095162 | |
| Gene (Homo sapiens) | Unc5D | GenBank | NCBI: 137970 Gene Cards: GC08P035235 | |
| Gene (Homo sapiens) | Dyrk1A | GenBank | NCBI: 1859 Gene Cards: GC21P037365 | |
| Gene (Homo sapiens) | DCC | GenBank | NCBI: 1630 Gene Cards: GC18P052340 | |
| Gene (Homo sapiens) | Neo1 | GenBank | NCBI: 4756 Gene Cards: GC15P073051 | |
| Gene (Homo sapiens) | DSCAM | GenBank | NCBI: 1826 Gene Cards :GC21M040010 | |
| Strain, strain background (Escherichia coli) | Endura competent cells | Biosearch Technologies | 60242-2 | Electrocompetent cells |
| Strain, strain background (Escherichia coli) | NEB10 beta | New England Biolabs | C3019H | High efficiency chemically Competent cells |
| Cell line (Homo-sapiens) | OVCAR8Ovarian cancer cell line | This paper | RRID:CVCL 1629 | Ovarian cancer cell line maintained in T.Shepherd lab |
| Cell line (Homo-sapiens) | OVCAR3Ovarian cancer cell line | ATCC | HTB-161 | |
| Cell line (Homo-sapiens) | TOV1946 ovarian cancer cell line | This paper | RRID:CVCL 4062 | Ovarian cancer cell line maintained in R. Rottapel lab |
| Cell line (Homo-sapiens) | iOvCa147Ovarian cancer cells | This paper | Primary ovarian cancer cell line maintained in T. Shepherd lab | |
| Cell line (Homo-sapiens) | OVCAR4Ovarian cancer cell line | Millipore-Sigma | SCC258 | |
| Cell line (Homo-sapiens) | COV318Ovarian cancer cell line | Millipore-Sigma | 07071903-1VL | Primary ovarian cancer cell line |
| Cell line (Homo sapiens) | Hek293T | ATCC | CRL-3216 RRID:CVCL_0063 | Human embryonic kidney cells |
| Biological sample (Homo sapiens) | EOC96Ovarian cancer cells | London Health Sciences Centre | Fresh isolate from patient ascites | |
| Biological sample (Homo sapiens) | EOC101Ovarian cancer cells | London Health Sciences Centre | Fresh isolate from patient ascites | |
| Biological sample (Homo sapiens) | EOC526Ovarian cancer cells | London Health Sciences Centre | Fresh isolate from patient ascites | |
| Antibody | Phosphor-p44/42 MAPK (Erk)Rabbit Monoclonalantibody | Cell Signaling technology | #4370 RRID:AB_2315112 | WB 1:1000 |
| Antibody | p44/42 MAPK(Erk) Rabbit Monoclonalantibody | Cell Signaling technology | #4695 RRID:AB_390779 | WB 1:1000 |
| Antibody | Phosphor-p38MAPKRabbit Monoclonalantibody | Cell Signaling technology | #4511 RRID:AB_2139682 | WB 1:1000 |
| Antibody | p38 MAPKRabbit Monoclonalantibody | Cell Signaling technology | #9215 RRID:AB_331762 | WB 1:1000 |
| Antibody | a-TubulinRabbit Monoclonalantibody | Cell Signaling Technology | #2125 RRID:AB_2619646 | WB 1:1000 |
| Antibody | NTN1Rabbit Monoclonalantibody | Abcam | #Ab126729 RRID:AB_11131145 | 1:1000 for Western blots1:150 for IHC |
| Antibody | p130(RBL2) Rabbit polyclonal | Santa Cruz | Discontinued RRID:AB_632093 | 1:1000 for Western Blots1:150 for IHC |
| Antibody | Ki67Rabbit Monoclonalantibody | Cell Signaling Technology | #12202 RRID:AB_2620142 | 1:500 for IHC |
| Antibody | Ki67Rabbit Monoclonalantibody | Abcam | #ab16667 RRID:AB_302459 | 1:1000 for Western blot |
| Antibody | p53Rabbit Monoclonalantibody | Cell Signaling Technology | #2527 RRID:AB_10695803 | 1:120 for IHC |
| Antibody | EpCAMRabbit Monoclonalantibody | Cell Signaling Technology | #93790 RRID:AB_2800214 | 1:150 for IHC |
| Antibody | Myc (9E10)Mouse Monoclonalantibody | Santa Cruz | #sc-40 RRID:AB_627268 | WB 1:1000 |
| Antibody | NTN3 | Gift from P. Mehlan lab | Not commercially available | 1:1000 for Western Blot1:500 for IHC |
| Antibody | Cytokeratin 7/8Mouse Monoclonalantibody | Zeta Corporation | RRID:AB_11162687 Discontinued | 1:250 for IHC |
| Antibody | Phosphor Tau(S404)Rabbit Monoclonalantibody | Cell Siganling Technology | #20194 RRID:AB_2798837 | WB 1:1000 |
| Antibody | TauRabbit Monoclonalantibody | Cell Signaling Technology | #46687 RRID:AB_2783844 | WB 1:1000 |
| Antibody | DYRK1ARabbit Polyclonalantibody | Cell Signaling Technology | #2771 RRID:AB_915851 | WB 1:1000 |
| Antibody | b-ActinRabbit Polyclonalantibody | Millipore Sigma | #A2066 RRID:AB_476693 | WB 1:1000 |
| Antibody | Goat anti-mouse IgG Biotinylated | Jackson ImmunoResearch | #115-067-003 RRID:AB_2338586 | IHC 1:500 |
| Antibody | Goat anti-rabbit IgG Biotinylated | Vector Laboratories | #BP-9100-50 | IHC 1:500 |
| Recombinant DNA reagent | Human GeCKO Lentiviral sgRNA library V2LentiGuide Puro | Addgene | #1000000049 | Human whole genome CRISPR library |
| Recombinant DNA reagent | Lenti-CAS9-Blast | Addgene | #52962 RRID:Addgene_52962 | Lentiviral vector for delivery of CAS9 |
| Recombinant DNA reagent | pLentiCRISPR-Puro-V2 | Addgene | #52961 RRID:Addgene_52961 | Lentiviral vector for delivery of sgRNA |
| Recombinant DNA reagent | pSPCAS9-(BB)2A-GFP(pX458) | Addgene | #48138 RRID:Addgene_48138 | Vector for delivery of sgRNA |
| Recombinant DNA reagent | NTN1-FC-His | Addgene | #72104 RRID:Addgene_72104 | Plasmid carrying full length Murine NTN1 fused to the FC portion of IgG |
| Recombinant DNA reagent | NTN3-FC-His | Addgene | #72105 RRID:Addgene_72105 | Plasmid carrying full length Murine NTN3 fused to the FC portion of IgG |
| Recombinant DNA reagent | FutdTW | Addgene | #22478 RRID:Addgene_22478 | Lentiviral expression vector |
| Recombinant DNA reagent | FutdTW NTN1 | This paper | Lentiviral vector expressing full length Murine NTN1 | |
| Recombinant DNA reagent | FutdTW NTN3 | This paper | Lentiviral vector expressing full length Murine NTN3 | |
| Recombinant DNA reagent | pcDNA3.1myc/His A | ThermoFisher | V80020 | Mammalian expression vector |
| Recombinant DNA reagent | pcDNA3.1myc/His ANTN1 | This paper | Mammalian expression vectorfor the expression of NTN1 under G418 selection | |
| Recombinant DNA reagent | pcDNA3.1myc/His ANTN3 | This paper | Mammalian expression vector for the expression of NTN3 under G418 selection | |
| Recombinant DNA reagent | LentiCRISPRV2NTN1_A | This paper | Lentiviral vector for the delivery of, sgRNA targeting human NTN1 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN1_B | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN1 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN1_C | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN1 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN3_1 | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN3 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN3_2 | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN3 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN3_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN3 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN4_A | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN4 | |
| recombinant DNA reagent | LentiCRISPRV2NTN4_B | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN4 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN5_A | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN5 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN5_B | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN5 | |
| Recombinant DNA reagent | LentiCRISPRV2NTN5_C | This paper | Lentiviral vector for the delivery of sgRNA targeting human NTN5 | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5A_1 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5A | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5A_2 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5A | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5A_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5A | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5B_1 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5B | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5B_2 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5B | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5B_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5B | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5C_1 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5C | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5C_2 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5C | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5C_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5C | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5D_1 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5D | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5D_2 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5D | |
| Recombinant DNA reagent | LentiCRISPRV2Unc5D_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Unc5D | |
| Recombinant DNA reagent | LentiCRISPRV2DCC_1 | This paper | Lentiviral vector for the delivery of sgRNA targeting human DCC | |
| Recombinant DNA reagent | LentiCRISPRV2DCC_2 | This paper | Lentiviral vector for the delivery of sgRNA targeting human DCC | |
| Recombinant DNA reagent | LentiCRISPRV2DCC_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human DCC | |
| Recombinant DNA reagent | LentiCRISPRV2Neo1_1 | This paper | Lentiviral vector for the deliveryof sgRNA targeting human Neo1 | |
| Recombinant DNA reagent | LentiCRISPRV2Neo1_2 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Neo1 | |
| Recombinant DNA reagent | LentiCRISPRV2Neo1_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human Neo1 | |
| Recombinant DNA reagent | LentiCRISPRV2DSCAM_1 | This paper | Lentiviral vector for the deliveryof sgRNA targeting human DSCAM | |
| Recombinant DNA reagent | LentiCRISPRV2DSCAM_2 | This paper | Lentiviral vector for the deliveryof sgRNA targeting human DSCAM | |
| Recombinant DNA reagent | LentiCRISPRV2DSCAM_3 | This paper | Lentiviral vector for the delivery of sgRNA targeting human DSCAM | |
| Sequence-based reagent | NTN1-AsgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-GCAGTCGTCGG CGGCGCTAC-3’ |
| Sequence-based reagent | NTN3-AsgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-CGACTGTCCGG CCGCCGCAG-3’ |
| Sequence-based reagent | NTN4-AsgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-CACATTAACGT CGAAGTGAC-3’ |
| Sequence-based reagent | NTN5-AsgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-ATCGTAGCATG GGTCCGCAG-3’ |
| Sequence-based reagent | Unc5A-1sgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-CTGTGCTGCG CTCGATCACG-3’ |
| Sequence-based reagent | Unc5B-1sgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-CGTACAGGCG ATGCGGACGT-3’ |
| Sequence-based reagent | Unc5C-1sgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-TCCCTTCAGG TGGTCGACAC-3’ |
| Sequence-based reagent | Unc5D-1sgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-CTTACAGGCTA TGCGCACAG-3’ |
| Sequence-based reagent | DCC-1sgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-GACTTCCTCG CCTCGTAACC-3’ |
| Sequence-based reagent | Neo1-1sgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-CGGGCTTTAT CGCTGCGTAG-3’ |
| Sequence-based reagent | DSCAM-1sgRNA | GeCKO genomic CRISPRLibrary V2 | CRISPR guide | 5’-ATCGTAGATC TCCTCGCCCG-3’ |
| Peptide, recombinant protein | Recombinant Human Netrin-1 | R and D Systems | #6419-N1 | 500 ng/ml |
| Commercial assay or kit | Monarch DNA Gel Extraction Kit | New England Biolabs | #T1020 | |
| Commercial assay or kit | Monarch total RNA Miniprep Kit | New England Biolabs | #T2010 | |
| Commercial assay or kit | Monarch PCR DNA Cleanup kit | New England Biolabs | #T1030 | |
| Commercial assay or kit | NEBNext Ultra II Q5 Master Mix | New England Biolabs | #MO544 | |
| Commercial assay or kit | Monarch Plasmid MiniPrep Kit | New England Biolabs | #T1010 | |
| Commercial assay or kit | DNeasy Blood and Tissue DNA isolation kit | Qiagen | #69504 | |
| Commercial assay or kit | NextSeq 75 Cycle NG sequencing kit | Illumina | #20024906 | |
| Commercial assay or kit | iScript cDNA Synthesis Kit | Bio-Rad | #1708890 | |
| Commercial assay or kit | iQ SYBR Green Supermix | Bio-Rad | #1708882 | |
| Commercial assay or kit | RNA 6000 Nano Kit | Agilent Technologies | #5067-1511 | |
| Commercial assay or kit | ScriptSeq Complete Gold Kit | Illumina | #BEP1206 | |
| Commercial assay or kit | NextSeq 500Mid-Output150 cycles | Illumina | #20024907 | |
| Chemical compound, drug | Trematinib | Selleckchem | #S4484 | |
| Chemical compound, drug | Polybrene | Santa Cruz | #sc-134220 | 8 mg/ml |
| Chemical compound, drug | Crystal Violet | Millipore Sigma | #C6158 | 0.50% |
| Chemical compound, drug | Protease inhibitor cocktail | Cell Signaling Technology | #5871S | |
| Chemical compound, drug | Phospahtase inhibitor cocktail | Cell Signaling Technology | #5870S | |
| Software, algorithm | Graphpad | Prism | RRID:SCR_002798 | |
| Software, algorithm | Image Lab | Bio-Rad | RRID:SCR_014210 | |
| Software, algorithm | Aperio ImageScope | Leica | RRID:SCR_020993 | |
| Software, algorithm | SAMTOOLS | htslib.org | RRID:SCR_002105 | |
| Software, algorithm | Illumina Sequencing HUB (BaseSpace) | Illumina | RRID:SCR_011881 | |
| Software, algorithm | Cutadapt | code.google.com/p/cutadapt/ | RRID:SCR_011841 | |
| Software, algorithm | Bowtie 2 | bowtie-bio.sourceforge.net/bowtie2/index.shtml | RRID:SCR_016368 | |
| Software, algorithm | ConsensusPathDB | Cpdb.molgen.mpg.de | RRID:SCR_002231 | |
| Software, algorithm | Reactome | https://reactome.org/ | RRID:SCR_003485 | |
| Software, algorithm | MAGeCK | https://sourceforge.net/projects/mageck/ | ||
| Software, algorithm | STAR2.6.1a | https://code.google.com/archive/p/rna-star/ | RRID:SCR_004463 | |
| Software, algorithm | DESeq2 | http://bioconducter.org/packages/release/bioc/html/DESeq2/html | RRID:SCR_015687 | |
| Software, algorithm | CBioPortal | cbioportal.org | RRID:SCR_014555 | |
| Software, algorithm | The Cancer Genome Atlas (TCGA) | cancergenome.nih.gov | RRID:SCR_003193 | |
| Software, algorithm | R 3.6.2 | https://www.r-project.org/ | RRID:SCR_001905 | |
| Software, algorithm | BEAVR | https://github.com/developerpiru/BEAVR; https://hub.docker.com/r/pirunthan/beavr; Perampalam and Dick, 2020 | BMC Bioinformatics. 2020 May 29;21(1):221. | |
| Other | Streptavidin HRP | Vector Laboratories | #SA-5704-100 | Secondary for IHCSee Methods, Histology and immunohisto-chemistry |
| Other | ImmPACT DAB | Vector Laboratories | #SK-4105 | Chromophore for IHCSee Methods, Histology and immunohisto-chemistry |
| Other | VectaMount | Vector Laboratories | #H-5700-60 | Mounting materialfor IHC glass slides See Methods, Histology and immunohisto-chemistry |
| Other | Hematoxylin | Millipore Sigma | #HHS32-1L | Counterstain reagentFor IHCSee Methods, Histology and immunohisto-chemistry |
| Other | Eosin Y | Millipore Sigma | #E4009 | Common stain in conjunction with hematoxylinfor IHC of tissue See Methods, Histologyand immunohisto-chemistry |
| Other | Gibson Assembly Mastermix | New England Biolabs | #E2611 | See “Methods” Generation of Knockout Lines |
| Other | Dynabeads | Thermo-Fisher | #10003D | See Methods, Dyrk1AIP kinase assay |
| Other | BsmB1 | New England Biolabs | #R0739 | Restriction endonucleasefor construction of CRISPR constructs |
| Other | EcoRV | New England Biolabs | #R0195 | Restriction endonuclease for construction of pcDNA NTN1. See “Methods”Generation of Overexpression cell lines |
| Other | XbaI | New England Biolabs | #R0145 | Restriction endonuclease for construction of pcDNA NTN1. See “Methods”Generation of Overexpression cell lines |
| Other | NotI | New England Biolabs | #R3189 | Restriction endonuclease for construction of pcDNA NTN3. See “Methods”Generation of Overexpression cell lines |
| Other | BamH1 | New England Biolabs | #R0136 | Restriction endonuclease for construction of FutdTWNTN1 and NTN3. See “Methods”Generation of Overexpression cell lines |
| Other | EcoRI | New England Biolabs | #R0101 | Restriction endonucleasefor construction of FutdTWNTN1 and NTN3. See “Methods”Generation of Overexpression cell lines |
| Sequence-based reagent | v2.1-F1 | TKO libraryProtocols Addgene#90294 | PCR Primer for gen omic amplification of CRISPR guides http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-GAGGGCCTATT TCCCATGATTC-3’ |
| Sequence-based reagent | v2.1-R1 | TKO libraryProtocols Addgene#90294 | PCR Primer for ge nomic amplification of CRISPR guides http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-GTTGCGAAAAA GAACGTTCACGG-3’ |
| Sequence-based reagent | D501 -F | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATGATACGGCGACCACC GAGATCTACACTATAG CCTACACTCTTTCCCTACA CGACGCTCTTCCGAT CTTTGTGG AAAGGACGAAACACCG-3’ |
| Sequence-based reagent | D502-F | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATGATACGGCGACCA CCGAGATCTACACATAGA GGCACACTCTTTCCCTA CACGACGCTCTTCCGAT CTTTGTGGA AAGGACGAAACACCG-3’ |
| Sequence-based reagent | D503-F | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATGATACGGCGACCA CCGAGATCTACACCCT ATCCTACACTCTTTCCCT ACACGACGCTCTTCCG ATCTTTGTGGA AAGGACGAAACACCG-3’ |
| Sequence-based reagent | D504-F | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATGATACGGCGACCACC GAGATCTACACGGCT CTGAACACTCTTTCCCTA CACGACGCTCTTCCG ATCTTTGTGG AAAGGACGAAACACCG-3’ |
| Sequence-based reagent | D505-F | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATGATACGGCGACCA CCGAGATCTACACAGGC GAAGACACTCTTTCCCTA CACGACGCTCTTCCGAT CTTTGTGGA AAGGACGAAACACCG-3’ |
| Sequence-based reagent | D506-F | TKO library Protocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATGATACGGCGACCA CCGAGATCTACACTAA TCTTAACACTCTTTCCCTA CACGACGCTCTTCCG ATCTTTGTG GAAAGGACGAAACACCG-3’ |
| Sequence-based reagent | D701-R | TKO library Protocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-CAAGCAGAAGACGG CATACGAGATCGAGTAA TGTGACTGGAGTTCAGA CGTGTGCTCTTCCGAT CTACTTGCTAT TTCTAGCTCTAAAAC-3’ |
| Sequence-based reagent | D702-R | TKO library Protocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATGATACGGCGACCA CCGAGATCTACACATAGA GGCACACTCTTTCCCTA CACGACGCTCTTCCGAT CTTTGTGG AAAGGACGAAACACCG-3’ |
| Sequence-based reagent | D704-R | TKO library Protocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-CAAGCAGAAGACGGCA TACGAGATGGAATCTCGT GACTGGAGTTCAGACGT GTGCTCTTCCGATCTAC TTGCTATTTCTA GCTCTAAAAC-3’ |
| Sequence-based reagent | D705-R | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-CAAGCAGAAGACGGCAT ACGAGATTTCTGAATGTG ACTGGAGTTCAGACGTGTG CTCTTCCGATCTACTTG CTATTTCTA GCTCTAAAAC-3’ |
| Sequence-based reagent | D706-R | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-CAAGCAGAAGACGGCAT ACGAGATACGAATTCGT GACTGGAGTTCAGACGTGTGCTCT TCCGATCTAC TTGCTATTTCTAGCTCTAAAAC-3’ |
| Sequence-based reagent | D707-R | TKO libraryProtocols Addgene#90294 | Indexing Primer http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-CAAGCAGAAGACGG CATACGAGATAGCTTCAGGT GACTGGAGTTCAGAC GTGTGCTCTTCCGATCTAC TTGCTATTTCTAG CTCTAAAAC-3’ |
| Sequence-based reagent | Dyrk1A sgRNA-A | This paper | CRISPR guide | 5'-CTCACTTAT CTTCTTGTAGG-3' |
| Sequence-based reagent | Dyrk1A sgRNA-B | This paper | CRISPR guide | 5'-GCAACGTG GGATTATGGATT-3' |
| Sequence-based reagent | NTN1-BsgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-ACCCGTCAC GCCGTCCTTGC-3’ |
| Sequence-based reagent | NTN1-CsgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5'-TATCGGCCA CGATGCCGCTC-3' |
| Sequence-based reagent | NTN3-BsgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GGTCTCGAT AGAAGCCCTCC-3’ |
| Sequence-based reagent | NTN3-CsgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-ACCTGCAAC CGCTGCGCGCC-3’ |
| Sequence-based reagent | NTN4-BsgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-CGCAGGTC ACGATAGAAGCC-3’ |
| Sequence-based reagent | NTN5-BsgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GCCGCCCG TCCCATCGAGAC-3’ |
| Sequence-based reagent | NTN5-CsgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GGCCTGAC CTGCAACCGCTG-3’ |
| Sequence-based reagent | Unc5A-2sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-CGCCCG CGGCCATGGCCGTC-3’ |
| Sequence-based reagent | Unc5A-3sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GTCCTCGCCG CTTGGCTCCG-3’ |
| Sequence-based reagent | Unc5B-2sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-TTCACAAT GTAGGCGTCCTG-3’ |
| Sequence-based reagent | Unc5B-3sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-CCTGTGTGA CGTGGTCGTTC-3’ |
| Sequence-based reagent | Unc5C-2sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-TGAGATTT CGCGCCAGCAAG-3’ |
| Sequence-based reagent | Unc5C-3sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-CAATGCGC ACATACGCCTTC-3’ |
| Sequence-based reagent | Unc5D-2sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GCGCTTA CCTCGGGCAGCCG-3’ |
| Sequence-based reagent | Unc5D-3sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-CAGAGACGT GCTCGTTCTGA-3’ |
| Sequence-based reagent | DCC-2sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-AAATTCCAA TGTCCCCCGGT-3’ |
| Sequence-based reagent | DCC-3sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GCAGATCA GCCGACTCCAAC-3’ |
| Sequence-based reagent | Neo1-2sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GAACCTTC CTCAGTTTATGC-3’ |
| Sequence-based reagent | Neo1-3sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-TGTTTCCCA CGTAACAGTGA-3’ |
| Sequence-based reagent | DSCAM-2sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-AGTGATGTAC GCCTCCACCG-3’ |
| Sequence-based reagent | DSCAM-3sgRNA | GeCKO genomicCRISPRLibrary V2Addgene#1000000049 | CRISPR guide | 5’-GGAGCCCTAT ACAGTCCGTG-3’ |
| Sequence-based reagent | Dyrk1A Exon2 F | PCR prime | 5’-GGTTTCACCT GGTTTGGGGA-3’ | |
| Sequence-based reagent | Dyrk1A Exon2 R | PCR prime | 5’-TCCGTGGG CAAGAAACTTT-3’ | |
| Sequence-based reagent | Unc5A-FqRCR primer | PCR primer | 5’-GCTGAGGCGC TAAAGCCGCCCTC-3’ | |
| Sequence-based reagent | Unc5A-RqRCR primer | PCR primer | 5’-ACCTGCTGCCT TGAGACATTAATGC-3’ | |
| Sequence-based reagent | Unc5B-FqPCR primer | PCR primer | 5’-CCCGCCACA CAGATCTACTT-3’ | |
| Sequence-based reagent | Unc5B-RqPCR primer | PCR primer | 5’-CAGTAATCC TCCAGCCCAAA-3’ | |
| Sequence-based reagent | Unc5C_1-FqPCR primer | PCR primer | 5’-GCAAATTGCTG GCTAAATATCAGGAA-3’ | |
| Sequence-based reagent | Unc5C_1-RqPCR primer | PCR primer | 5’-GCTCCACTGTGT TCAGGCTAAATCTT-3’ | |
| Sequence-based reagent | Unc5C_2-FqPCR primer | PCR primer | 5’-AATTGATCC CGTTGAAGATCGG-3’ | |
| Sequence-based reagent | Unc5C_2-RqPCR primer | PCR primer | 5’-TGACAGTGG CAGTTGTACTTTT-3’ | |
| Sequence-based reagent | Unc5D_1-FqPCR primer | PCR primer | 5’-CAAGAGCAA CCCTATTGCACT-3’ | |
| Sequence-based reagent | Unc5D_1-RqPCR primer | PCR primer | 5’-CTCGTTCTG ATGGACCCACT-3’ | |
| Sequence-based reagent | Unc5D_2-FqPCR primer | PCR primer | 5’-CAAGAGCAA CCCTATTGCACT-3’ | |
| Sequence-based reagent | Unc5D_2_RqPCR primer | PCR primer | 5’-AAGCCCTTCC CGAATCCATC-3’ | |
| Sequence-based reagent | NTN1-FqPCR primer | PCR primer | 5’-TGCAAGAAGG ACTATGCCGTC-3’ | |
| Sequence-based reagent | NTN1-RqPCR primer | PCR primer | 5’-GCTCGTGCCC TGCTTATACAC-3’ | |
| Sequence-based reagent | NTN3-FqPCR primer | PCR primer | 5’-TGCAAGCCCT TCTACTGCGACA-3’ | |
| Sequence-based reagent | NTN3-RqPCR primer | PCR primer | 5’-CAGTCGGTA CAGCTCCATGTTG-3’ | |
| Sequence-based reagent | NTN4-FqPCR primer | PCR primer | 5'-CAGAAGGACAG TATTGCCAGAGG-3' | |
| Sequence-based reagent | NTN4-RqPCR primer | PCR primer | 5'-GCAGAAGGTC ACTGAGTTGGCA-5' | |
| Sequence-based reagent | NTN5-FqPCRprimer | PCR primer | 5'-CTTGCCACTA CTCCTGGTGCTT-3' | |
| Sequence-based reagent | NTN5-RqPCR primer | PCR primer | 5'-AGTACCTC CGAAGGCTCATGTG-3' | |
| Sequence-based reagent | DCC_1-FqPCR primer | PCR primer | 5’-GACTTTACCAAT GTGAGGCATCT-3’ | |
| Sequence-based reagent | DCC_1-RqPCR primer | PCR primer | 5’-GGTCCTGCT ACTGCAACTTTT-3’ | |
| Sequence-based reagent | DCC_2-FqPCRprimer | PCR primer | 5’-GAGACACA GTGCTACTCAAGTG-3’ | |
| Sequence-based reagent | DCC_2-RqPCR primer | PCR primer | 5’-GGAGTCAGG TCTTGTTGGTTCTT-3’ | |
| Sequence-based reagent | DSCAM_1-FqPCR primer | PCR primer | 5’-TTGCGGTCT TCAAGTGCATTA-3’ | |
| Sequence-based reagent | DSCAM_1-RqPCR primer | PCR primer | 5’-TGCAGCGGTAGTTATACAATCCA-3’ | |
| Sequence-based reagent | DSCAM_2-FqPCR primer | PCR primer | 5’-ATCAGACCCAGCGAACTCAG-5’ | |
| Sequence-based reagent | DSCAM_2-RqPCR primer | PCR primer | 5’-CCAGCGGTAATCTGGCTCAG-3’ | |
| Sequence-based reagent | Neo-1-FqPCR primer | PCR primer | 5’-GTCACTGAGACCTTGGTAAGCG-3’ | |
| Sequence-based reagent | Neo-1-RqPCR primer | PCR primer | 5’-TCAGCAGACAGCCAGTCAGTTG-3’ | |
| Sequence-based reagent | GAPDH-FqPCR primer | PCR primer | 5'-CGGAGTCAACGGATTTGGTCGTAT-3' | |
| Sequence-based reagent | GAPDH-RqPCR primer | PCR primer | 5'-AGCCTTCTCCATGGTGGTGAAGAC-3' | |
| Sequence-based reagent | NTN1-F cloning primer_1 | Cloning primer | 5’- GCTATCGATATCCCAAACG CCACCATGATGCGCGC TGTGTGGGAGGCGCTG -3’ | |
| Sequence-based reagent | NTN1-Rcloning primer | Cloning primer | 5’- GCTATCCTCGAGGGCCTTCTTGCAC TTGCCCTTCTTCTCCCG -3’ | |
| Sequence-based reagent | NTN3-Fcloning primer_1 | Cloning primer | 5’-GCTAGCGCGGCCGCCACCATGC CTGGCTGGCCCTGG-3’ | |
| Sequence-based reagent | NTN3-Rcloning primer | Cloning primer | 5’- ACGCGTGAATTCTTATCAA CCGGTATGCATATTCA GATCCTCTTCTGAGAT -3’ | |
| Sequence-based reagent | NTN1-Fcloning primer_2 | Cloning primer | 5’-CACTGTAGATCTCCAAACGC CACCATGATGCGCGCTG TGTGGGAGGCGCTG-3’ | |
| Sequence-based reagent | NTN3-FCloning primer_2 | Cloning primer | 5’-CTCGAGATCTGCGGCCGCCACC ATGCCTGGCTGGCCCTGG-3’ | |
| Sequence-based reagent | NTN1_NTN3Cloning reverse primer | Cloning primer | 5’-ACGCGTGAATTCTTATCAA CCGGTATGCATATTCA GATCCTCTTCTGAGAT-3’ | |
| Sequence-based reagent | LacZ control CRISPR guide | CRISPR guide | 5’-CCCGAATCTCTA TCGTGCGG-3’ | |
| Sequence-based reagent | PSMD1-1 | Fitness gene CRISPR guide http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-TGTGCGCTACG GAGCTGCAA-3’ | |
| Sequence-based reagent | PSMD1-5 | Fitness gene CRISPR guide http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-ACCAGAGCCAC AATAAGCCA-3’ | |
| Sequence-based reagent | EIF3D | Fitness gene CRISPR guide http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-ACCGACTTCAAC TGAAGAGTCTCG-3’ | |
| Sequence-based reagent | PSMB2 | Fitness gene CRISPR guide http://dx.doi.org/10.1016/j.cell.2015.11.015 | 5’-AATATTGTCCAGA TGAAGGA-3’ |





