Phosphorylation bar-coding of free fatty acid receptor 2 is generated in a tissue-specific manner
Figures
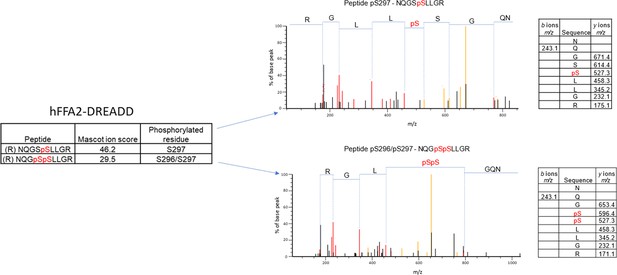
Mass spectrometry analysis of hFFAR2-DREADD-eYFP identifies basal phosphorylation of Ser297 and agonist-promoted phosphorylation of Ser296.
Mass spectrometry analysis was conducted on samples isolated from Flp-In T-REx 293 cells in which expression of hFFAR2-DREADD-eYFP had been induced. Experiments were performed on vehicle and 4-methoxy-3-methyl-benzoic acid (MOMBA)-treated (100 μM, 5 min) cells as detailed in Materials and methods. LC-MS/MS identified Ser297 as being phosphorylated constitutively, and Ser296/297 as being phosphorylated by sorbic acid or MOMBA. Composite outcomes of a series of independent experiments are combined. Fragmentation tables associated with phosphorylated peptides are shown. Phosphorylated residues are highlighted in red.
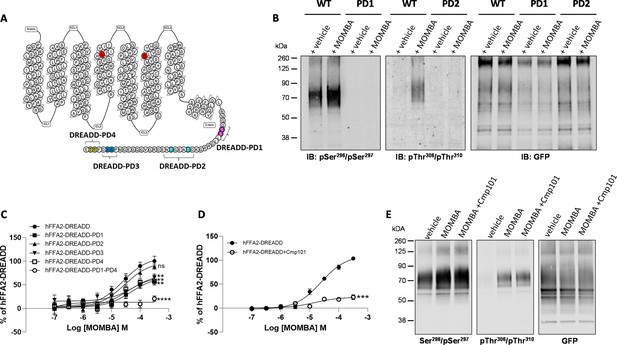
Characteristics of putative pSer296/pSer297 and pThr306/pThr310 hFFAR2-antisera and the effect of potential phospho-acceptor site mutations on agonist-induced arrestin-3 interactions.
The primary amino acid sequence of hFFAR2 is shown (A). Residues altered to generate the DREADD variant are in red (Cys141Gly, His242Gln). Phospho-deficient (PD) hFFAR2-DREADD variants were generated by replacing serine 296 and serine 297 (purple, hFFAR2-DREADD-PD1), threonine 306 and threonine 310 (light blue, hFFAR2-DREADD-PD2), serine 324 and serine 325 (dark blue, hFFAR2-DREADD-PD3) or threonine 328 and 329 (yellow, hFFAR2-DREADD-PD4) with alanine. In addition, hFFAR2-DREADD-PD1-4 was generated by combining all these alterations. (B) The ability of putative pSer296/pSer297 and pThr306/Thr310 antisera to identify wild-type and either PD1 or PD2 forms of hFFAR2-DREADD with and without treatment of cells expressing the various forms with 4-methoxy-3-methyl-benzoic acid (MOMBA) is shown and, as a control, anti-GFP immunoblotting of equivalent samples is illustrated. (C) The ability of varying concentrations of MOMBA to promote interaction of arrestin-3 with hFFAR2-DREADD and each of the DREADD-PD mutants is illustrated. Each of the DREADD-PD variants, except hFFAR2-DREADD-PD2 (ns), were less effective in promoting interactions in response to MOMBA (**p<0.01, ****p<0.0001). (D) The effect of the GRK2/3 inhibitor compound 101 on the capacity of MOMBA to promote recruitment of arrestin-3 to wild-type hFFAR2-DREADD is shown (***p<0.001). Significance in C and D were assessed by one-way ANOVA followed by Dunnett’s multiple comparisons test. (E) The effect of compound 101 on detection of hFFAR2-DREADD-eYFP by each of pSer296/pSer297, pThr306/Thr310, and anti-GFP antisera is shown. Data are representative (B, E) or show means ± SEM (C, D) of at least three independent experiments.
-
Figure 2—source data 1
The ability of putative pSer296/pSer297 antisera to identify wild-type and PD1 forms of hFFAR2-DREADD.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig2-data1-v1.zip
-
Figure 2—source data 2
The ability of putative and pThr306/Thr310 antisera to identify wild-type and PD2 forms of hFFAR2-DREADD.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig2-data2-v1.zip
-
Figure 2—source data 3
GFP control for phosphor-antisera to identify wild-type and mutant forms of hFFAR2-DREADD.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig2-data3-v1.zip
-
Figure 2—source data 4
4-Methoxy-3-methyl-benzoic acid (MOMBA) promotes interaction of arrestin-3 with hFFAR2-DREADD and each of the DREADD-PD mutants.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig2-data4-v1.xlsx
-
Figure 2—source data 5
The effect of compound 101 on detection of hFFAR2-DREADD-eYFP by pSer296/pSer297.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig2-data5-v1.zip
-
Figure 2—source data 6
The effect of compound 101 on detection of hFFAR2-DREADD-eYFP by pThr306/Thr310.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig2-data6-v1.zip
-
Figure 2—source data 7
GFP control for phosphor-antisera to identify the effect of compound 101.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig2-data7-v1.zip
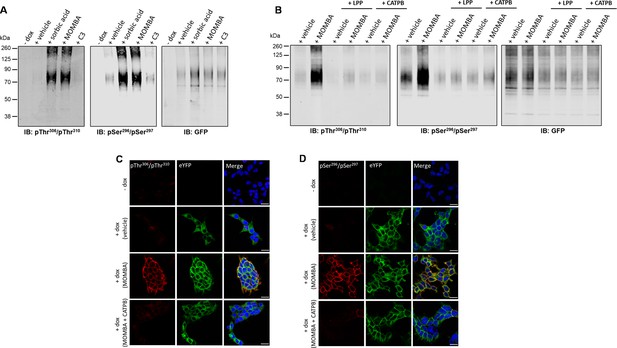
Agonist-induced detection of hFFAR2-DREADD with putative pSer296/pSer297 and pThr306/pThr310 antisera reflects receptor activation, receptor phosphorylation, and can be detected in situ.
The ability of the pSer296/pSer297, pThr306/pThr310 hFFAR2, and, as a control GFP, antisera to identify hFFAR2-DREADD-eYFP after induction to express the receptor construct and then treatment of cells with vehicle, 4-methoxy-3-methyl-benzoic acid (MOMBA), sorbic acid (each 100 µM), or propionate (C3) (2 mM) is shown. In the ‘-dox’ lanes receptor expression was not induced. (B) As in (A) except that after cell treatment with vehicle or MOMBA, immune-enriched samples were treated with lambda protein phosphatase (LPP) or, rather than treatment with MOMBA, cells were treated with a combination of MOMBA and the hFFAR2 inverse agonist ((S)-3-(2-3-chlorophenyl)acetamido)-4-(4-(trifluoromethyl)phenyl)butanoic acid (CATPB) (10 µM, 30 min pre-treatment). (C, D) Cells harboring hFFAR2-DREADD-eYFP and grown on glass coverslips were either untreated (- dox) or induced to express hFFAR2-DREADD-eYFP. The induced cell samples were then exposed to vehicle, MOMBA (100 µM), or a combination of MOMBA (100 µM) and CATPB (10 µM) for 5 min. Fixed cells were then treated with anti-pThr306/pThr310 (C) or anti-pSer296/pSer297 (D) (red, Alexa Fluor 647) or imaged to detect eYFP (green). DAPI was added to detect DNA and highlight cell nuclei (blue). Scale bar = 20 μm.
-
Figure 3—source data 1
Agonist-induced detection of hFFAR2-DREADD with pTHr306/pThr310antisera.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig3-data1-v1.zip
-
Figure 3—source data 2
Agonist-induced detection of hFFAR2-DREADD with pSer296/pSer297 antisera.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig3-data2-v1.zip
-
Figure 3—source data 3
GFP control for agonist-induced detection of hFFAR2-DREADD.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig3-data3-v1.zip
-
Figure 3—source data 4
Activation and inhibition of hFFAR2-DREADD with pThr306/pThr310 antisera.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig3-data4-v1.zip
-
Figure 3—source data 5
Activation and inhibition of hFFAR2-DREADD with pSer296/pSer297 antisera.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig3-data5-v1.zip
-
Figure 3—source data 6
GFP control for activation and inhibition of hFFAR2-DREADD.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig3-data6-v1.zip
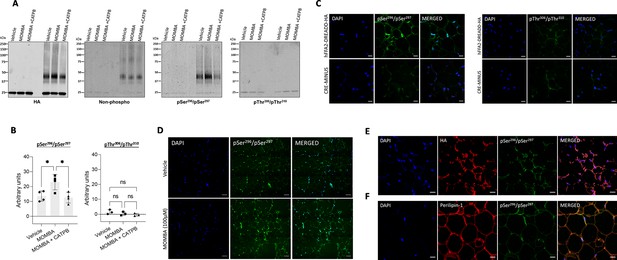
In white adipose tissue residues Ser296/Ser297 of hFFAR2-DREADD-HA but not Thr306/Thr310 become phosphorylated in response to 4-methoxy-3-methyl-benzoic acid (MOMBA).
White adipose tissue dissected from hFFAR2-DREADD-HA and CRE-MINUS mice was treated with either vehicle, 100 µM MOMBA, or 100 µM MOMBA+10 µM ((S)-3-(2-3-chlorophenyl)acetamido)-4-(4-(trifluoromethyl)phenyl)butanoic acid (CATPB). (A) Lysates were prepared and solubilised. hFFAR2-DREADD-HA was immunoprecipitated using an anti-HA monoclonal antibody and following SDS-PAGE immunoblotted to detect HA, non-phosphorylated hFFAR2-DREADD-HA, and pSer296/pSer297 or pThr306/pThr310hFFAR2-DREADD-HA. A representative experiment is shown. (B) Quantification of pSer296/pSer297 (left) and pThr306/pThr310 immunoblots (right) phosphorylation (means ± SEM) in experiments using tissue from different mice, *p<0.05, ns: not significant. Significance was assessed by one-way ANOVA, followed by Tukey’s multiple comparisons test (n=4). (C) Tissue samples from hFFAR2-DREADD-HA (top panel) and CRE-MINUS (bottom panel) mice that were treated with MOMBA were immunostained to detect pSer296/pSer297 (left panels), pThr306/pThr310 (right panels) and counterstained with DAPI (blue). Scale bars = 20 µm. (D) Comparison of pSer296/pSer297 staining of samples from hFFAR2-DREADD-HA-expressing mice vehicle treated (top panels) or treated with MOMBA (bottom panels) (scale bar = 50 µm). (E, F) Tissue sections from hFFAR2-DREADD-HA-expressing mice immunostained with pSer296/pSer297 (green) and anti-HA (red) (E) to detect the receptor expression or F with anti-perilipin-1 (red) to identify adipocytes. Merged images are shown to the right. Scale bars = 20 µm.
-
Figure 4—source data 1
hFFAR2-DREADD-HA expression in white adipose tissue.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig4-data1-v1.zip
-
Figure 4—source data 2
pSer296/pSer297 detects phosphorylation of hFFAR2-DREADD-HA in white adipose tissue.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig4-data2-v1.zip
-
Figure 4—source data 3
pThr306/pThr310 is not phosphorylated in white adipose tissue.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig4-data3-v1.zip
-
Figure 4—source data 4
Quantification of pSer296/pSer297 and pThr306/pThr310 immunoblots in white adipose tissue.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig4-data4-v1.xlsx
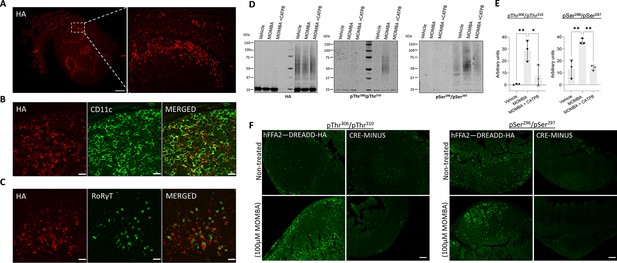
hFFAR2-DREADD-HA becomes phosphorylated at Thr306/Thr310 in addition to Ser296/Ser297 in immune cells from Peyer’s patches.
Peyer’s patches isolated from hFFAR2-DREADD-HA-expressing mice were immunostained with anti-HA (red) to detect receptor expression. Images were acquired with ×20 (left panel) and ×63 (right panel) objectives (scale bar = 200 µm) (A). Tissue sections were counterstained with (B) anti-CD11c as a marker of dendritic cells, monocytes, and/or macrophages or (C) RORγT to detect type-III innate lymphoid cells (scale bar = 20 µm). Isolated Peyer’s patches and mesenteric lymph nodes from CRE-MINUS and hFFAR2-DREADD-HA mice were exposed to either vehicle, 100 µM 4-methoxy-3-methyl-benzoic acid (MOMBA) or 100 µM MOMBA+10 µM ((S)-3-(2-3-chlorophenyl)acetamido)-4-(4-(trifluoromethyl)phenyl)butanoic acid (CATPB). (D) Following lysate preparation, immunoprecipitation and SDS-PAGE samples were probed to detect HA, pThr306/pThr310, or pSer296/pSer297. (E) Quantification of pThr306/pThr310 (left) and pSer296/pSer297 immunoblots (right) hFFAR2-DREADD-HA (means ± SEM), *p<0.05, **p<0.01. Significance was assessed by one-way ANOVA, followed by Tukey’s multiple comparisons test (n=3). (F) Treated tissue sections were also used in immunohistochemical studies, employing either pThr306/pThr310 (left panels) or pSer296/pSer297 (right panels) (scale bars = 100 µm).
-
Figure 5—source data 1
hFFAR2-DREADD-HA expression in Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig5-data1-v1.zip
-
Figure 5—source data 2
pThr306/pThr310 detects agonist-dependent phosphorylation in Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig5-data2-v1.zip
-
Figure 5—source data 3
pSer296/pSer297 detects both constitutive and agonist-dependent phosphorylation in Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig5-data3-v1.zip
-
Figure 5—source data 4
Quantification of pSer296/pSer297 and pThr306/pThr310 immunoblots in Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig5-data4-v1.xlsx
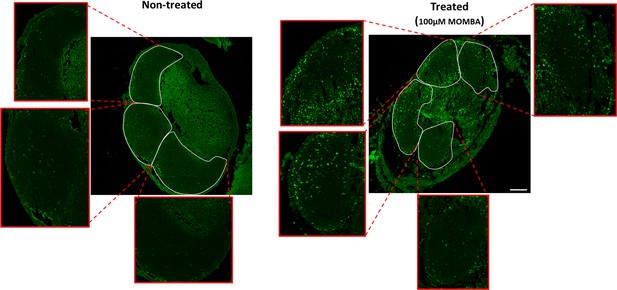
hFFAR2-DREADD-HA becomes phosphorylated at Thr306/Thr310 in immune cells within Peyer’s patches.
Experiments akin to those of Figure 5F were conducted using the pThr306/pThr310 hFFAR2 antiserum. The figure illustrates 4-methoxy-3-methyl-benzoic acid (MOMBA)-induced phosphorylation of pThr306/pThr310 in immune cells within Peyer’s patches of hFFAR2-DREADD-HA-expressing mice. Peyer’s patches were either treated with vehicle (left panel) or MOMBA (100 µM) (right panel). Each lymphoid nodule has been expanded to show detailed phosphorylation inside each nodule.

4-Methoxy-3-methyl-benzoic acid (MOMBA) promotes limited phosphorylation of both Ser296/Ser297 and Thr306/Thr310 in hFFAR2-DREADD-HA in lower gut enteroendocrine cells.
(A, B) Colonic tissue isolated from hFFAR2-DREADD-HA (top panels) or CRE-MINUS (bottom panels) mice treated with either vehicle (A) or 100 µM MOMBA (B). Following fixation tissue sections were immunostained with pThr306/pThr310 and counterstained with DAPI (scale bar = 100 µm). In the merged images, the box is expanded in the right-hand panels. (C) Lysates prepared from tissue samples treated as noted were analysed by probing immunoblots with anti-HA, anti-pThr306/pThr310, or anti-pSer296/pSer297. Representative examples are shown.
-
Figure 6—source data 1
hFFAR2-DREADD-HA expression in colonic epithelium.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig6-data1-v1.zip
-
Figure 6—source data 2
pThr306/pThr310 detects phosphorylation in colonic crypts.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig6-data2-v1.zip
-
Figure 6—source data 3
pSer296/pSer297 detects phosphorylation in colonic crypts.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig6-data3-v1.zip

Propionate regulates phosphorylation of hFFAR2-eYFP: in vitro studies.
Flp-In T-REx 293 cells habouring hFFAR2-eYFP were induced to express the receptor construct (+dox) or not (-dox) and the induced cells were then treated with propionate (C3, 2 mM, 5 min) or vehicle. (A) Cell lysates were resolved by SDS-PAGE and then immunoblotted with anti-pSer296/pSer297 hFFAR2, anti-pThr306/pThr310 hFFAR2, or anti-GFP. (B) Cells induced to express hFFAR2-eYFP were treated with C3 (2 mM, 5 min) or vehicle. Where noted cells were pre-treated with the hFFAR2 antagonist (S)-3-(2-(3-chlorophenyl)acetamido)-4-(4-(trifluoromethyl)phenyl)butanoic acid (CATPB) (10 μM, 20 min before agonist addition). Lysates were then prepared and, where indicated, treated with lambda protein phosphatase (LPP). Following SDS-PAGE the samples were immunoblotted with anti-pThr306/pThr310 hFFAR2. (C, D) Cells were doxycycline induced (+dox) or not (-dox) and prepared for immunocytochemistry after treatment with C3 or vehicle and exposed to anti-pThr306/pThr310 hFFAR2 (C) or anti-pSer296/pSer297 hFFAR2 (D) (red) whilst direct imaging detected the presence of hFFAR2-eYFP (green). Merged images (right-hand panels) were also stained with DAPI (blue) to identify cell nuclei. Scale bars = 20 µm.
-
Figure 7—source data 1
The ability of putative pSer296/pSer297 antisera to identify phosphorylation of hFFAR2-eYFP.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig7-data1-v1.zip
-
Figure 7—source data 2
The ability of putative and pThr306/Thr310 antisera to phosphorylation of hFFAR2-eYFP.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig7-data2-v1.zip
-
Figure 7—source data 3
GFP control for phosphor-antisera to identify phosphorylation of hFFAR2-eYFP.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig7-data3-v1.zip
-
Figure 7—source data 4
The ability of pThr306/Thr310 antisera to detect phosphorylation and inhibition of hFFAR2-eYFP.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig7-data4-v1.zip
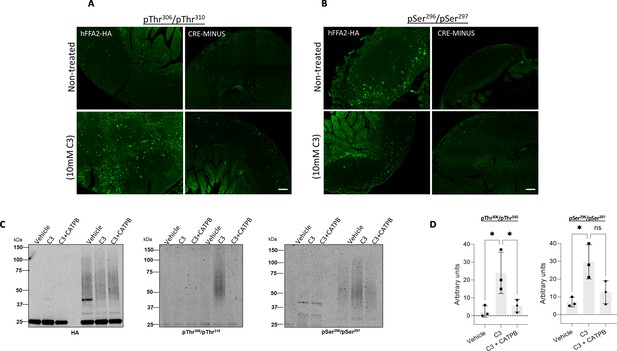
C3 induces phosphorylation of both pSer296/pSer297 and pThr306/pThr310 in Peyer’s patches from hFFAR2-HA-expressing mice.
Isolated Peyer’s patches and mesenteric lymph nodes from hFFAR2-HA and the corresponding CRE-MINUS mice were exposed to either vehicle or 10 mM C3 for 20 min. Tissue sections were used in immunohistochemical studies, employing either anti-pThr306/pThr310 (A) or anti-pSer296/pSer297 (B) (scale bars = 100 µm). (C) Lysates from Peyer’s patches isolated from hFFAR2-HA-expressing mice, or the corresponding CRE-MINUS mice, that had been treated with vehicle, C3 (10 mM, 20 min), or C3+(S)-3-(2-(3-chlorophenyl)acetamido)-4-(4-(trifluoromethyl)phenyl)butanoic acid (CATPB) (10 μM, 30 min before agonist) were immunoprecipitated with anti-HA as for the hFFAR2-DREADD-HA-expressing mice in Figure 5. Subsequent to SDS-PAGE samples such were probed to detect HA (C, left), anti-pThr306/pThr310(C, centre), or anti-pSer296/pSer297(C, right). hFFAR2-HA was detected as a broad smear of protein(s) with Mr centred close to 55 kDa. (D) Quantification of pThr306/pThr310 (left) and pSer296/pSer297 immunoblots (right) phosphorylation in experiments using tissue from three different mice (means ± SEM), *p<0.05, ns: not significant. Significance was assessed by one-way ANOVA, followed by Tukey’s multiple comparisons test (n=3).
-
Figure 8—source data 1
hFFAR2-HA expression in Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig8-data1-v1.zip
-
Figure 8—source data 2
pThr306/pThr310 detects agonist-dependent phosphorylation in hFFAR2-HA Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig8-data2-v1.zip
-
Figure 8—source data 3
pSer296/pSer297 detects agonist-dependent phosphorylation in hFFAR2-HA Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig8-data3-v1.zip
-
Figure 8—source data 4
Quantification of pSer296/pSer297 and pThr306/pThr310 immunoblots for hFFAR2-HA in Peyer’s patches.
- https://cdn.elifesciences.org/articles/91861/elife-91861-fig8-data4-v1.xlsx
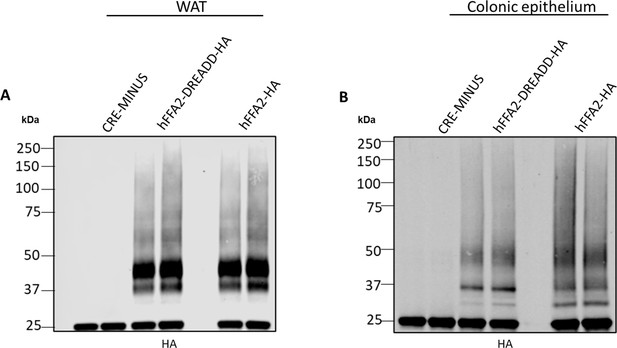
Tissues of transgenic mice express similar levels of hFFAR2-HA and hFFAR2-DREADD-HA.
White adipose (WAT) (left panel) and colonic epithelial (right panel) tissue was isolated from CRE-MINUS and both hFFAR2-DREADD-HA and hFFAR2-HA-expressing transgenic mice. Anti-HA immunoprecipitations were then performed, and samples resolved by SDS-PAGE followed by immunoblotting with anti-HA. For both tissues similar levels and patterns of the receptor proteins were detected from the hFFAR2-DREADD-HA and hFFAR2-HA-expressing mice, whilst these were absent in tissue from CRE-MINUS animals. A representative experiment of three is shown.
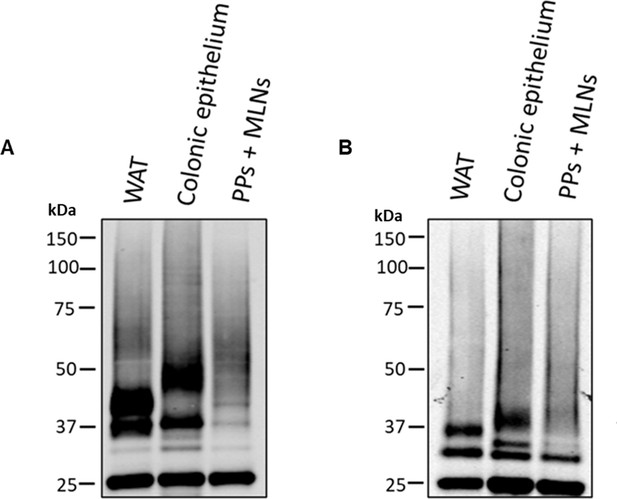
hFFAR2-DREADD-HA is present as multiple differentially N-glycosylated species in adipose tissue, immune, and colonic epithelial cells.
HA-immunoprecipitated samples from white adipose tissue (WAT), colonic epithelium, and Peyer’s patches and mesenteric lymph nodes (PP+MLNs) of hFFAR2-DREADD-HA-expressing mice were untreated (A) or treated with N-glycosidase F to remove N-linked carbohydrate (B). They were then resolved by SDS-PAGE and immunoblotted with anti-HA. A representative experiment of three is shown.





