Polysaccharides induce deep-sea Lentisphaerae strains to release chronic bacteriophages
Figures
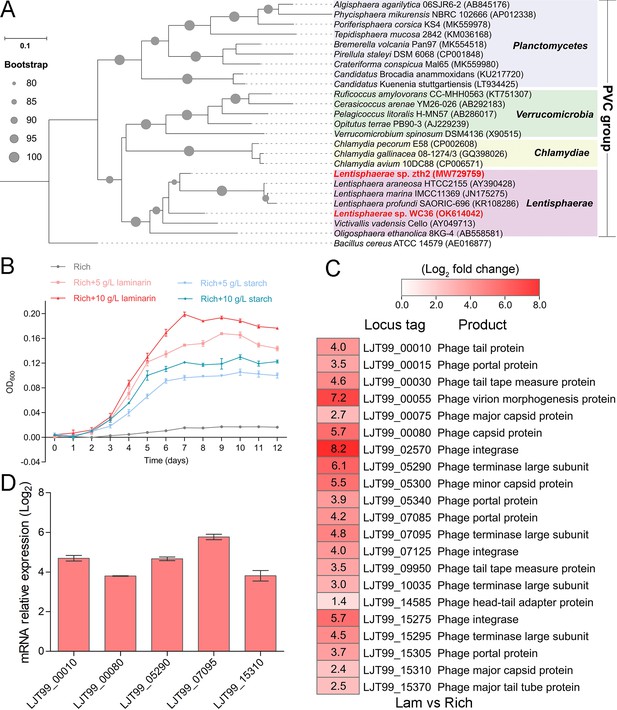
Polysaccharides promote the deep-sea Lentisphaerae strain WC36 growth and stimulate the expression of phage-associated genes.
(A) Maximum likelihood phylogenetic tree of 16S rRNA gene sequences from strain WC36, strain zth2, and some Planctomycetes–Verrucomicrobia–Chlamydia (PVC) group bacteria. Bacillus cereus ATCC 14579 was used as the outgroup. Bootstrap values (%) >80 are indicated at the base of each node with the gray dots (expressed as percentages of 1000 replications). The accession number is displayed behind corresponding strain. (B) Growth assays of strain WC36 cultivated in rich medium either without supplementation, with 5 or 10 g/l laminarin or with 5 or 10 g/l starch. (C) Transcriptomics-based heat map showing all upregulated genes encoding phage-associated proteins in strain WC36 cultured in rich medium supplemented with 10 g/l laminarin. ‘Rich’ indicates strain WC36 cultivated in rich medium; ‘Lam’ indicates strain WC36 cultivated in rich medium supplemented with 10 g/l laminarin. (D) Quantitative real-time polymerase chain reaction (qRT-PCR) detection of the expression of some genes encoding phage-associated proteins shown in panel C. The heat map was generated by Heml 1.0.3.3. The numbers in panel C represent multiple differences in gene expression (by taking log2 values).
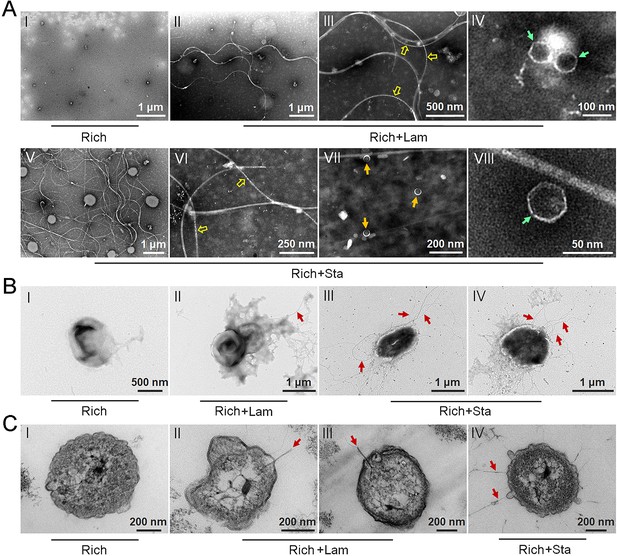
Polysaccharides induce the production of bacteriophages in Lentisphaerae strain WC36.
(A) Transmission electron microscopy (TEM) observation of phages extracted from the supernatant of WC36 cells cultured in rich medium supplemented with or without polysaccharide. Panel I shows the absence of phages in the supernatant of WC36 cells cultivated in rich medium. Panels II–IV show the morphology of phages present in the supernatant of WC36 cells cultivated in rich medium supplemented with 10 g/l laminarin. Yellow hollow arrows indicate the typical granular structure of filamentous phages, and green arrows indicate phage-like particles with other shapes. Panels V–VIII show the morphology of phages present in the supernatant of strain WC36 cells cultivated in rich medium supplemented with 10 g/l starch. Typical filamentous phages are indicated with yellow hollow arrows, and two other kinds of phage-like particles with different shapes are indicated by orange and green arrows, respectively. (B) TEM observation of strain WC36 cultured in rich medium supplemented with or without 10 g/l laminarin or 10 g/l starch. Panel I shows representative morphology of strain WC36 cultivated in rich medium. Panel II shows the morphology of strain WC36 cultivated in rich medium supplemented with 10 g/l laminarin. Panels III and IV show the morphology of strain WC36 cultivated in rich medium supplemented with 10 g/l starch. Red arrows indicate filamentous phages associated with bacterial cells. (C) TEM of an ultrathin section of strain WC36 cultured in rich medium supplemented with or without 10 g/l laminarin or 10 g/l starch. Panel I shows an ultrathin section of strain WC36 cultivated in rich medium; panels II and III show ultrathin sections of strain WC36 cultivated in rich medium supplemented with 10 g/l laminarin; panel IV shows an observation of the ultrathin section of strain WC36 cultivated in rich medium supplemented with 10 g/l starch. Red arrows indicate filamentous phages being released from or entering bacterial cells.
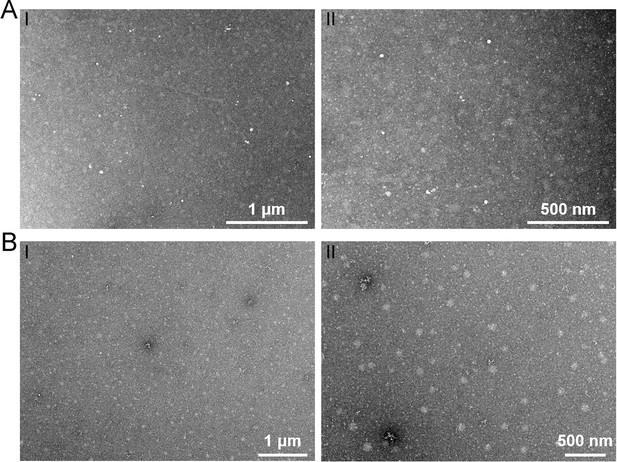
Transmission electron microscopy observation of phage particles in the supernatant of different mediums.
TEM observation of potential phage-like particles extracted from the rich medium supplemented with 10 g/l laminarin alone (A) or 10 g/l starch alone (B).
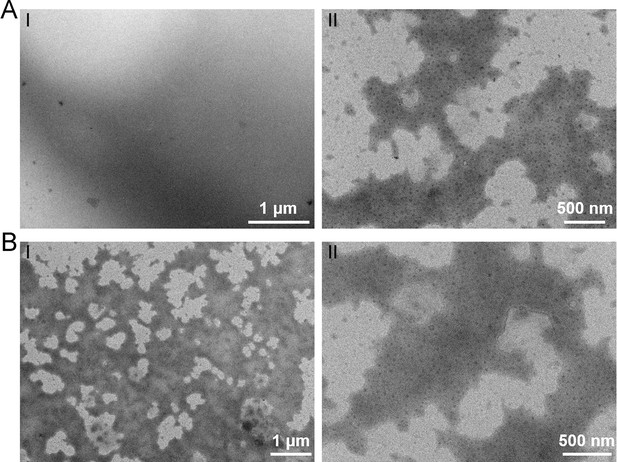
Transmission electron microscopy observation of different polysaccharides.
TEM observation of the rich medium supplemented with 10 g/l laminarin alone (A) or 10 g/l starch alone (B).
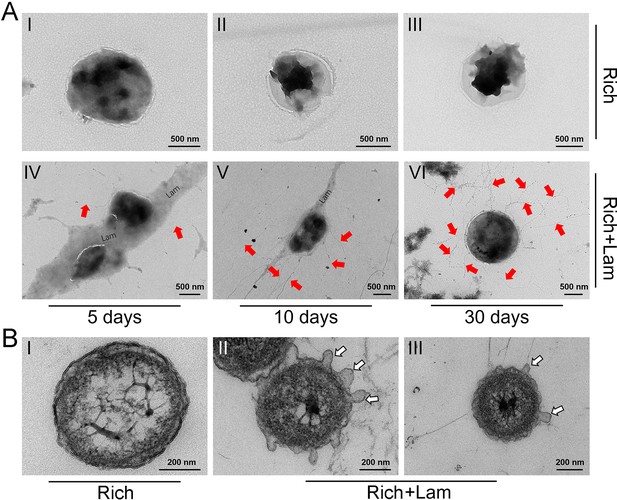
The morphology of Lentisphaerae strain WC36 and its released filamentous bacteriophages.
(A) Transmission electron microscopy (TEM) observation of the morphology of released filamentous bacteriophages and strain WC36 cultured in rich medium supplemented without (panels I–III) or with (panels IV–VI) 10 g/l laminarin for 5, 10, and 30 days, respectively. ‘Lam’ indicates the potential laminarin adhered to bacterial cells. The filamentous phages released from bacterial cells were pointed using red arrows. (B) TEM observation of an ultrathin section of strain WC36 cultured in rich medium supplemented without (panel I) or with (panels II and III) 10 g/l laminarin. White arrows indicate the extrusions or buddings around bacterial cells.
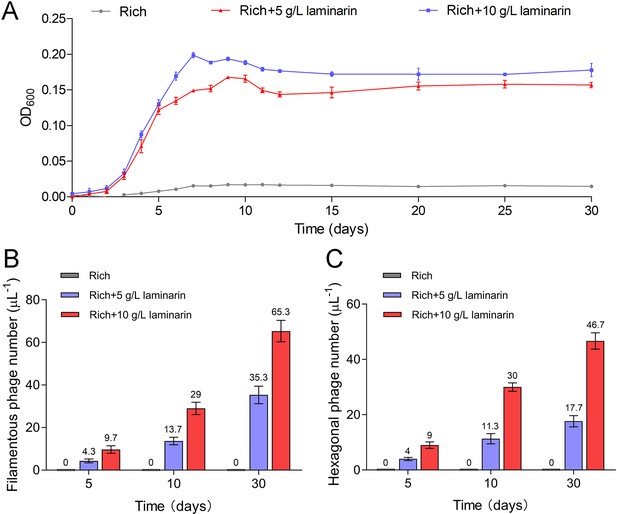
Bacteriophages induced from Lentisphaerae strain WC36 by polysaccharides are released via chronic manners.
(A) Growth curve of strain WC36 cultivated in either rich medium alone or rich medium supplemented with 5 or 10 g/l laminarin for 30 days. The number of filamentous phages (B) and hexagonal phages (C) extracted from 1 μl of the supernatant of strain WC36 cell cultured in rich medium supplemented with or without 5 or 10 g/l laminarin (for 5, 10, and 30 days). The average numbers are shown at the top of the bar charts.
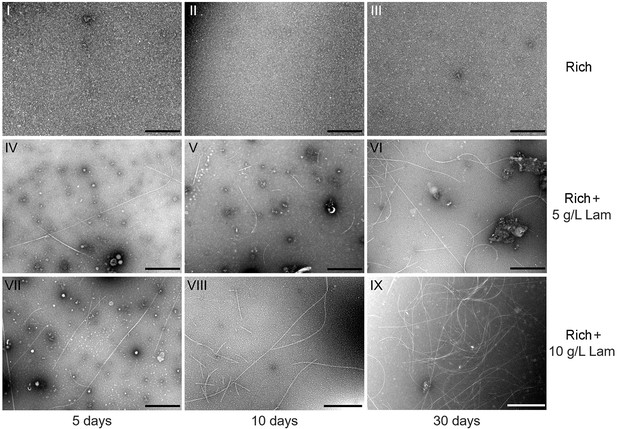
Transmission electron microscopy (TEM) observation of filamentous phages extracted from the supernatant of strain WC36 cell suspension cultured in rich medium supplemented with or without 5 or 10 g/l laminarin (for 5, 10, and 30 days).
Panels I–III show the absence of phages in the supernatant of cell strain WC36 cultivated in rich medium for 5, 10, and 30 days, respectively. Panels IV–VI show the morphology of filamentous phages present in the supernatant of cell strain WC36 cultivated in rich medium supplemented with 5 g/l laminarin for 5, 10, and 30 days, respectively. Panels VII–IX show the morphology of filamentous phages present in the supernatant of cell strain WC36 cultivated in rich medium supplemented with 10 g/l laminarin for 5, 10, and 30 days, respectively. Scale bars, 1 μm.
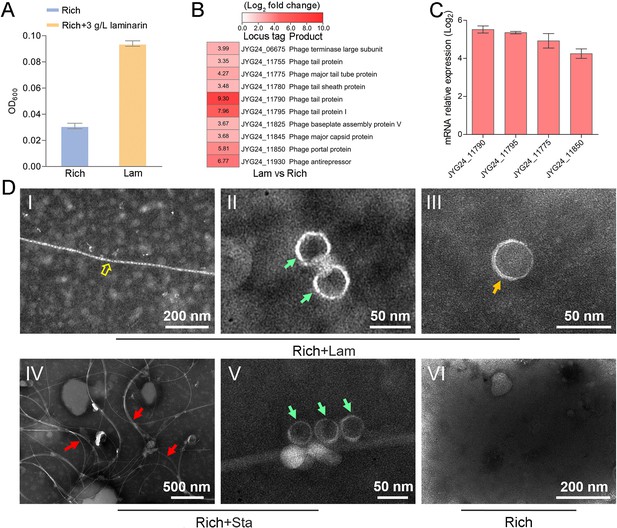
Polysaccharides promote the growth of deep-sea Lentisphaerae strain zth2 and induce the production of bacteriophages.
(A) Growth assays of strain zth2 cultivated in rich medium supplemented with or without 3 g/l laminarin for 4 days. (B) Transcriptomics-based heat map showing all upregulated genes encoding phage-associated proteins in strain zth2 cultured in rich medium supplemented with 3 g/l laminarin. (C) qRT-PCR detection of the expressions of some genes encoding phage-associated proteins shown in panel B. The numbers in panel C represent multiple differences in gene expression (by taking log2 values). (D) Transmission electron microscopy (TEM) observation of phages extracted from the supernatant of a cell suspension of strain zth2 cultured in the rich medium supplemented with or without polysaccharides. Panels I–III show the morphology of phages in the cell supernatant of strain zth2 cultivated in rich medium supplemented with 3 g/l laminarin. Typical filamentous phage is indicated with yellow hollow arrows, and two other kinds of phage-like particles with different shapes are indicated by orange and green arrows, respectively. Panels IV and V show the morphology of phages present in the cell supernatant of strain zth2 cultivated in rich medium supplemented with 3 g/l starch. Typical filamentous phages are indicated with red arrows, and other phage-like particles with different shapes are indicated by green arrows. Panel VI shows that no phages were observed in the cell supernatant of strain zth2 cultivated in rich medium. “Rich” indicates strain zth2 cultivated in rich medium; ‘Lam’ indicates strain zth2 cultivated in rich medium supplemented with 3 g/l laminarin.
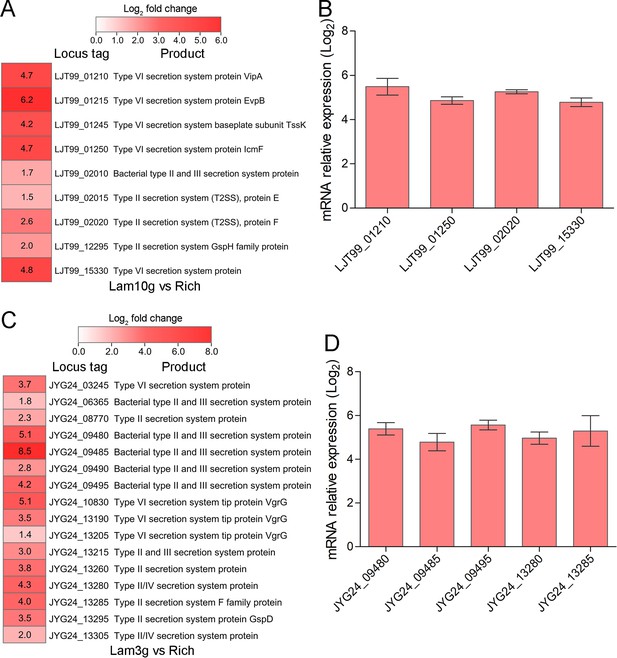
Transcriptome profiles and qRT-PCR analysis of the genes encoding secretion system-related proteins.
(A) Transcriptomics-based heat map showing all upregulated genes encoding secretion system-associated proteins in Lentisphaerae strain WC36. ‘Rich’ indicates strain WC36 cultivated in rich medium; ‘Lam’ indicates strain WC36 cultivated in rich medium supplemented with 10 g/l laminarin. (B) qRT-PCR detection of the expression of genes shown in panel A. (C) Transcriptomics-based heat map showing all upregulated genes encoding secretion system-associated proteins in strain zth2. ‘Rich’ indicates strain zth2 cultivated in rich medium; ‘Lam’ indicates strain zth2 cultivated in rich medium supplemented with 3 g/l laminarin. (D) qRT-PCR detection of the expression of genes shown in panel C. Three replicates were performed for each condition. The locus tag and its corresponding encoding product were shown with the heat map. The heat map is generated by Heml 1.0.3.3. The numbers in panels A and C represent multiple differences in gene expression (by taking log2 values).
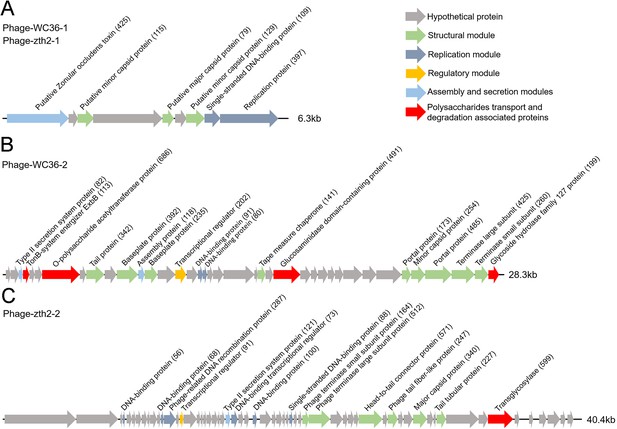
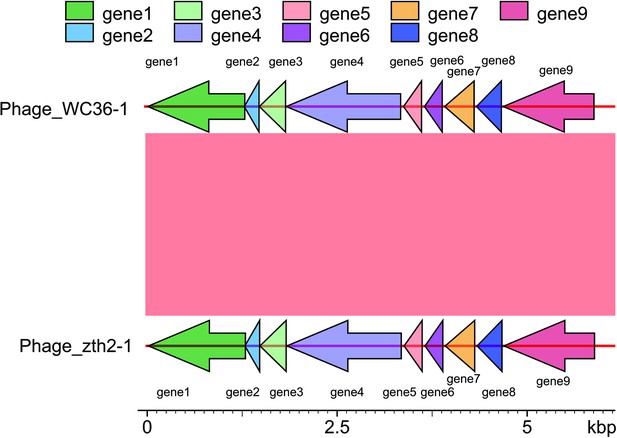
Genomic organization of bacteriophages released from Lentisphaerae strains WC36 and zth2 cultured in rich medium supplemented with polysaccharide.
(A) Schematic of the genomic composition of Phage-WC36-1 and Phage-zth2-1. (B) A diagram showing the genomic composition of Phage-WC36-2. (C) The genomic composition of Phage-zth2-2. Arrows represent different ORFs (open reading frames) and the direction of transcription. The main putative gene products of the phages are shown, and the numbers in brackets indicate the numbers of amino acids within each ORF. Hypothetical proteins are indicated with gray arrows, structural modules are indicated by green arrows, the replication module is indicated by blue-gray arrows, the regulatory module is indicated by golden arrows, the assembly and secretion modules are indicated by blue arrows and potential auxiliary metabolic genes (AMGs) encoding proteins associated with polysaccharide transport and degradation are indicated by red arrows. The size of the phage genomes is shown behind each gene cluster.
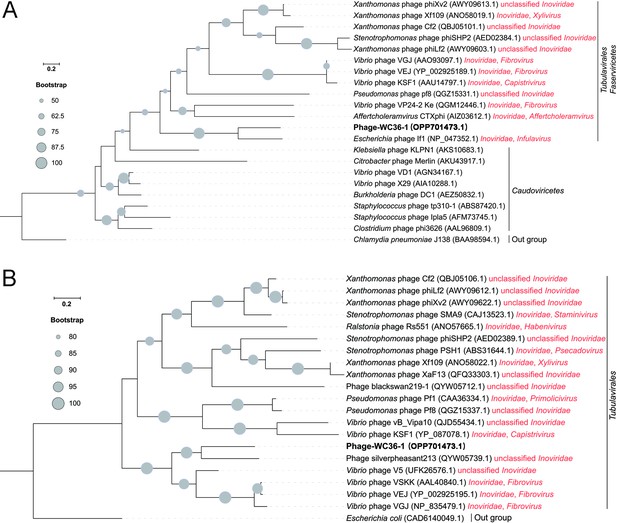
Phylogenetic analysis of Phage-WC36-1 and related phages.
(A) Maximum likelihood phylogenetic tree of single-stranded DNA-binding protein sequences from Phage-WC36-1 and some related phages. Chlamydia pneumoniae J138 was used as the outgroup. (B) Maximum likelihood phylogenetic tree of Zot protein sequences from Phage-WC36-1 and some related Tubulavirales phages. Escherichia coli was used as the outgroup. Bootstrap values (%) >50 (A) and >80 (B) are indicated at the base of each node with the gray dots (expressed as percentages of 1000 replications). The accession numbers of phages encoding single-stranded DNA-binding proteins and Zot proteins are given after the phage names.
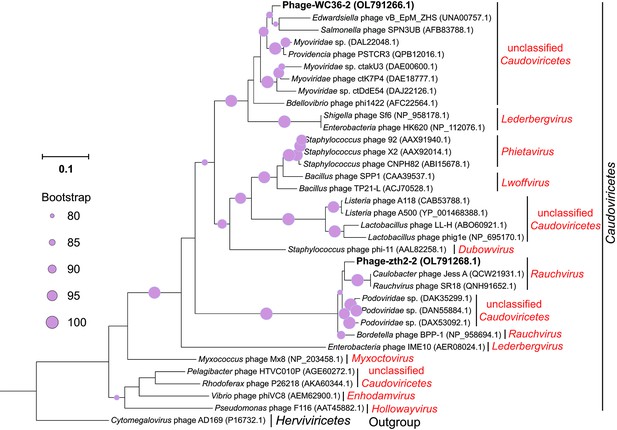
Maximum likelihood phylogenetic tree of terL protein sequences from Phage-WC36-2, Phage-zth2-2, and some related Caudoviricetes phages.
Cytomegalovirus phage AD169 was used as the outgroup. Bootstrap values (%) >80 are indicated at the base of each node with the gray dots (expressed as percentages of 1000 replications). The accession numbers of phages encoding terL proteins are given after the names of the phages.
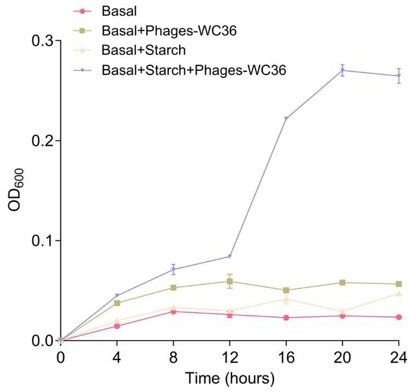
Growth curve and status of Pseudomonas stutzeri 273 cultivated in basal medium, basal medium supplemented with 20 μl/mL Phages-WC36, basal medium supplemented with 5 g/L starch, basal medium supplemented with 5 g/L starch and 20 μl/mL Phages-WC36.

Growth curve and status of Pseudomonas stutzeri 273 cultivated in basal medium, basal medium supplemented with 20 μl/mL Phages-WC36, basal medium supplemented with 5 g/L starch, basal medium supplemented with 5 g/L starch and 20 μl/mL Phages-WC36.

TEM observation of the supernatant of cells suspension of a Chloroflexi strain, a Tenericutes strain, a Proteobacteria strain and an Actinobacteria strain that cultivated in the rich medium supplemented with 10 g/L laminarin and 10 g/L starch.
No phage-like particles could be observed.
Tables
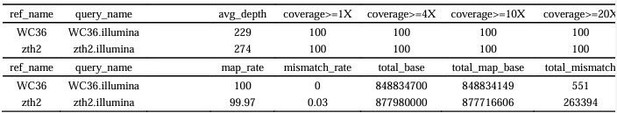
Sequencing depth and coverage statistics.
| ref_name | query_name | avg_depth | coverage >= 1X | coverage >= 4X | coverage >= 10X | coverage >= 20X |
|---|---|---|---|---|---|---|
| WC36 | WC36.illumina | 229 | 100 | 100 | 100 | 100 |
| zth2 | zth2.illumina | 274 | 100 | 100 | 100 | 100 |
| WC36 | WC36.illumina | 100 | 0 | 848834700 | 848834149 | 551 |
| zth2 | zth2.illumina | 99.97 | 0.03 | 877980000 | 877716606 | 263394 |
Additional files
-
Supplementary file 1
Annotations and fold-changes of the transcriptome data of Lentisphaerae strain WC36 cultured in rich medium supplemented with or without 10 g/l laminarin.
‘Rich’ indicates strain WC36 cultivated in rich medium; ‘Lam’ indicates strain WC36 cultivated in rich medium supplemented with 10 g/l laminarin.
- https://cdn.elifesciences.org/articles/92345/elife-92345-supp1-v1.xlsx
-
Supplementary file 2
Annotations of the transcriptome data of Lentisphaerae strain zth2 cultured in rich medium supplemented with or without 3 g/l laminarin.
‘Rich’ indicates strain zth2 cultivated in rich medium; ‘Lam’ indicates strain zth2 cultivated in rich medium supplemented with 3 g/l laminarin.
- https://cdn.elifesciences.org/articles/92345/elife-92345-supp2-v1.xlsx
-
Supplementary file 3
Annotations of the Phage-WC36-1 and Phage-zth2-1 genome.
- https://cdn.elifesciences.org/articles/92345/elife-92345-supp3-v1.xlsx
-
Supplementary file 4
Annotations of the Phage-WC36-2 genome.
- https://cdn.elifesciences.org/articles/92345/elife-92345-supp4-v1.xlsx
-
Supplementary file 5
Annotations of the Phage-zth2-2 genome.
- https://cdn.elifesciences.org/articles/92345/elife-92345-supp5-v1.xlsx
-
Supplementary file 6
Accession numbers of strain WC36, strain zth2, some Planctomycetes–Verrucomicrobia–Chlamydia (PVC) group bacteria, and Bacillus cereus ATCC 14579.
- https://cdn.elifesciences.org/articles/92345/elife-92345-supp6-v1.xlsx
-
Supplementary file 7
Primers used for qRT-PCR.
- https://cdn.elifesciences.org/articles/92345/elife-92345-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92345/elife-92345-mdarchecklist1-v1.docx