Divergent folding-mediated epistasis among unstable membrane protein variants
Figures
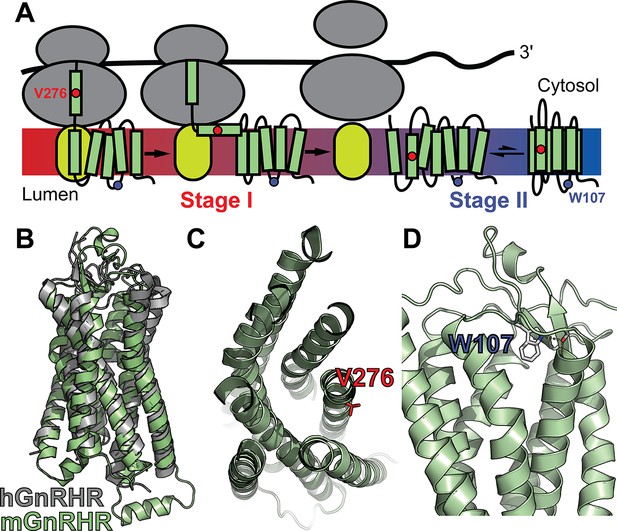
Mutagenic perturbation of GnRHR folding.
(A) A cartoon depicts the manner in which nascent polytopic membrane proteins are synthesized and folded at the endoplasmic reticulum (ER) membrane. The nascent protein first passes from a ribosome (gray) to a translocon (yellow), which facilitates its cotranslational membrane integration (stage I folding). Once the protein establishes its topology with respect to the membrane, it can fold into its native tertiary structure (stage II folding). (B) A homology model of M. musculus GnRHR (mGnRHR, green) is aligned with a crystal structure of H. sapiens GnRHR (hGnRHR, PDB 7BR3, gray). (C) A cross section of the mGnRHR model depicts the structural context of the V276 side chain (red). This side chain is located in transmembrane domain 6 and is exposed to the lipid bilayer. (D) A side view of the mGnRHR model depicts the structural context of the W107 side chain within extracellular loop 1. A hydrogen bond network between the W107 side chain indole, the backbone carbonyl oxygen of G110, and the backbone amide hydrogen from C114 is shown with dashes for reference.
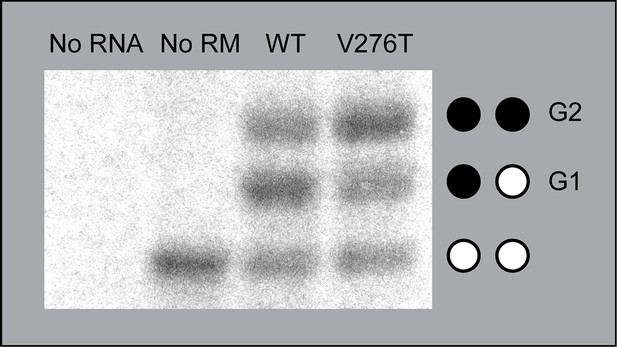
Membrane integration of mGnRHR TM domain 6 and V276T.
Chimeric Lep proteins containing either the wild type mGnRHR TM domain 6 or the V276T mutant, with the guest TM domain flanked by two glycosylation sites, were translated in vitro and analyzed by SDS-PAGE. Misintegration of the guest TM domain results in doubly glycosylated protein, while successful membrane integration results in singly glycosylated protein. Negative control translation reactions without RNA (first lane) and without rough microsomes (second lane) are included. The positions of the untargeted protein that is not associated with the membrane (no glycosylation), singly glycosylated form of the protein in which TMD6 is integrated into the membrane (G1), and doubly glycosylated form of the protein in which TMD6 is not integrated into the membrane (G2) are indicated for reference.
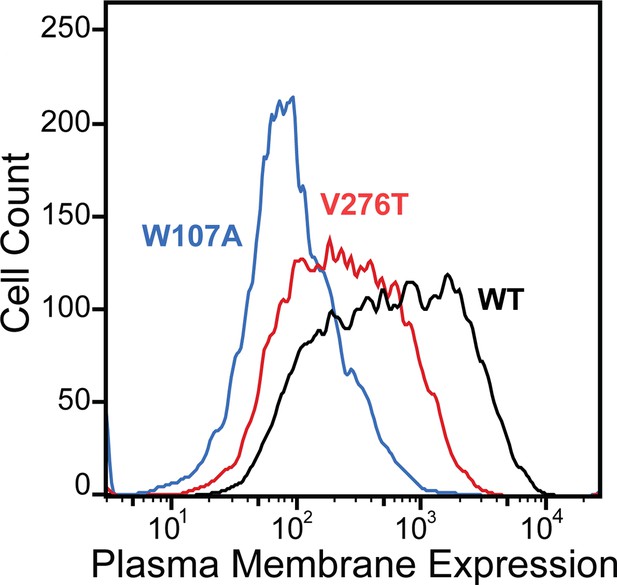
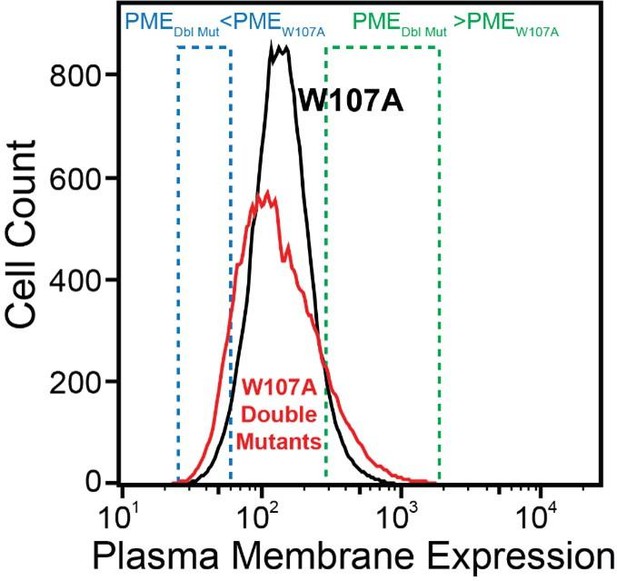
Plasma membrane expression of mGnRHR variants.
Surface immunostaining and flow cytometry were used to compare the plasma membrane expression of select mGnRHR variants expressed in HEK293T cells. A histogram from one representative biological replicate depicts the distribution of plasma membrane expression intensities among cells expressing WT (black), V276T (red), or W107A (blue) mGnRHR.
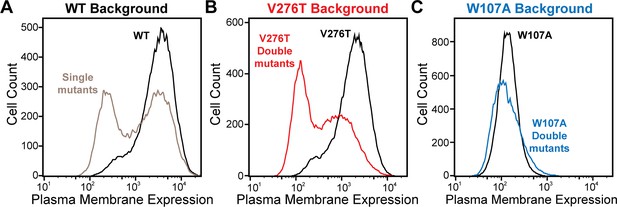
Plasma membrane expression of mGnRHR cellular libraries.
mGnRHR mutant libraries, as well as their respective controls (WT or V276T/W107A single mutants, black) were expressed in mammalian cells, and the amount of mGnRHR expressed at the plasma membrane was measured during fluorescence-activated cell sorting. Histograms show the distributions of plasma membrane expression for mGnRHR mutants in the (A) WT library (gray), (B) V276T library (red), and (C) W107A library (blue).
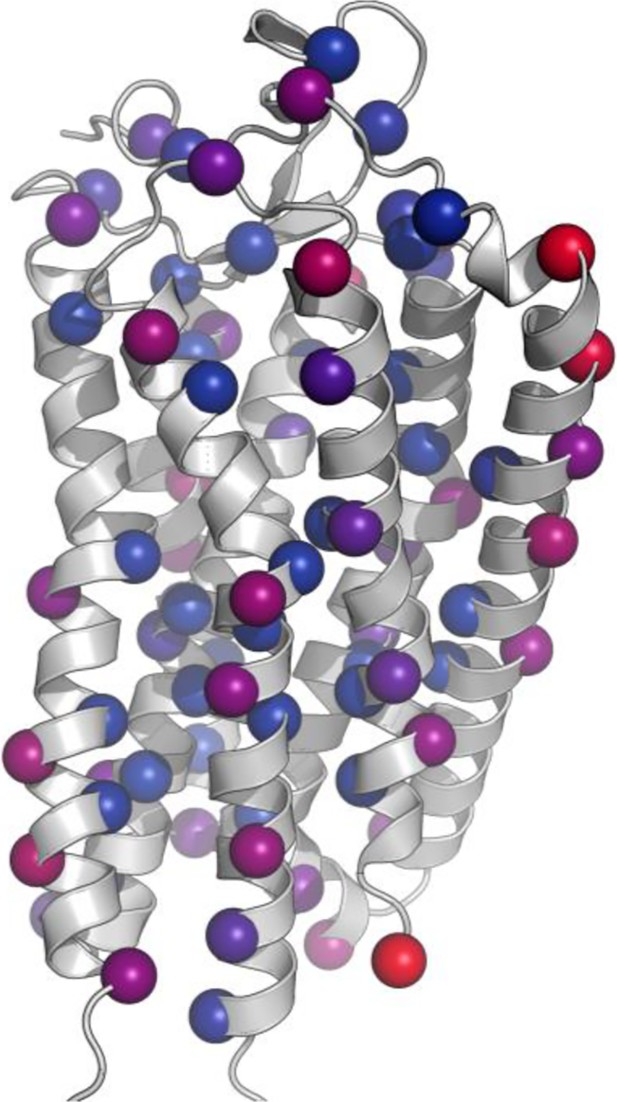
Structural context of mGnRHR library mutants.
Residues selected for mutation are displayed as spheres on the homology model of M. musculus GnRHR. Spheres are colored according to solvent accessible surface area of the residue, with red representing exposed residues and blue representing buried residues.
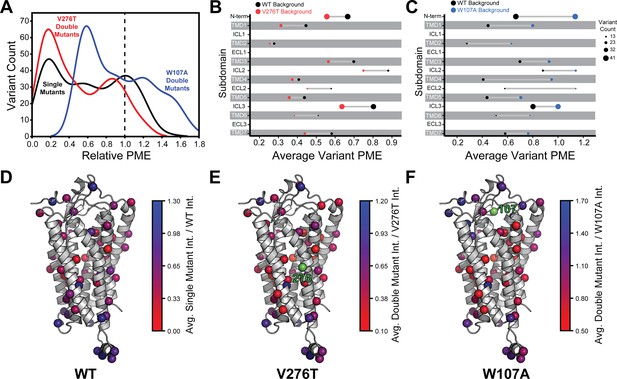
Relative plasma membrane expression of mGnRHR mutants.
(A) A histogram shows the distribution of plasma membrane expression measurements relative to their respective controls (WT or V276T/W107A single mutants) for the WT (black), V276T (red), and W107A (blue) mutational libraries. A dashed line at the normalized plasma membrane expression value of 1.0, representing no mutational effect relative to the control, is displayed for reference. (B, C) The average relative intensities across each domain are compared between the V276T (B, red) and W107A (C, blue) double mutant libraries and the single mutant library (black). The size of each dot represents the number of variants measured. (D–F) Average relative intensities for the (D) WT library, (E) V276T library, and (F) W107A library were projected onto a homology model of mGnRHR. Scores represent the average value from two biological replicates. Residues 276 and 107 are displayed in green for reference. Individual variant scores can be found in Supplementary file 4.
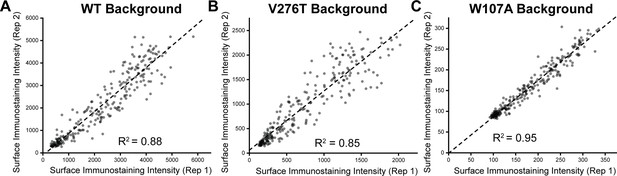
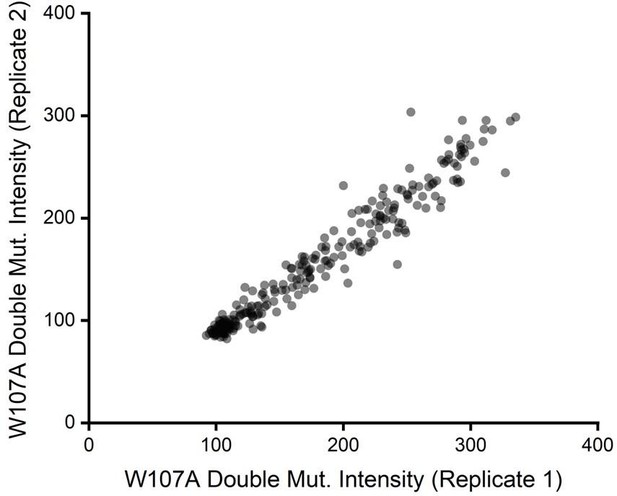
Correlation of deep mutational scanning (DMS) variant intensity values across two biological replicates.
The intensity values for the surface immunostaining of the 251 mutants scored the (A) WT, (B) V276T, and (C) W107A backgrounds determined from two independent biological replicates are plotted against one another. A linear fit (dashes) is shown for reference, along with the Pearson’s R2 value. These data demonstrate the precision of these measurements despite variations in the signal to noise that arises from variations in surface immunostaining intensity.
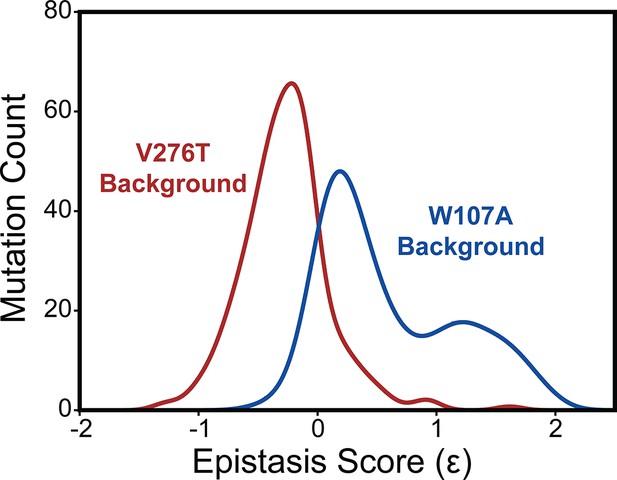
Epistasis in the mGnRHR double mutant libraries.
A histogram depicts the distribution of epistasis scores (Ɛ) for interactions the subset of 251 high-quality secondary mutations form with V276T (red) and W107A (blue).
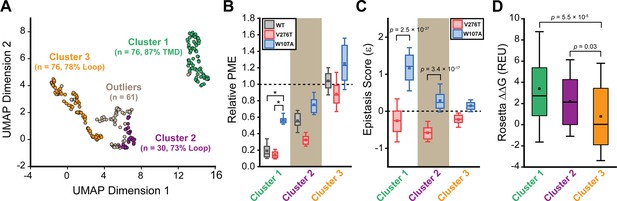
General trends in mGnRHR epistasis.
Trends associated with the observed pairwise epistasis within mGnRHR are identified using unsupervised learning. (A) Uniform manifold approximation projection (UMAP) was used to differentiate variants based on differences in their relative expression in the V276T, W107A, or WT background. Variants are projected onto an arbitrary two-dimensional coordinate based on the results and are colored according to whether they were assigned to cluster 1 (green), cluster 2 (purple), cluster 3 (orange), or were designated as outliers (gray) by HDBSCAN. The percentage of the mutations that fall within transmembrane domains (TMDs) or loops are shown for reference. (B) A box and whisker plot depicts the statistical distributions of relative plasma membrane expression (PME) values among variants within each cluster in the context of V276T (red), W107A (blue), or WT (gray) mGnRHR. Select clusters of variants that exhibit statistically different expression profiles according to a Mann–Whitney U-test are indicated (*p<0.001). A value of 1 corresponds to mutations that have no effect on the PME of mGnRHR in the indicated genetic background. (C) A box and whisker plot depicts the statistical distribution of epistasis scores associated with the interactions between the mutations within each cluster and either V276T (red) or W107A (blue). p-values for select Mann–Whitney U-tests comparing the interactions of these mutations with V276T and W107A are indicated. A value of 0 indicates that the effects of the two mutations are additive. (D) A box and whisker plot depicts the statistical distribution of Rosetta ΔΔG values among mutations within each cluster. p-values for select Mann–Whitney U-tests comparing the ΔΔG values across clusters are shown for reference. For panels (B–D), the edges of the boxes correspond to the 75th and 25th percentile values while the whiskers reflect the values of the 90th and 10th percentile. The central hash and square within the box represent the average and median values, respectively. These analyses were carried out on a subset of 243 variants with high-quality expression measurements with calculable Rosetta ΔΔG values.
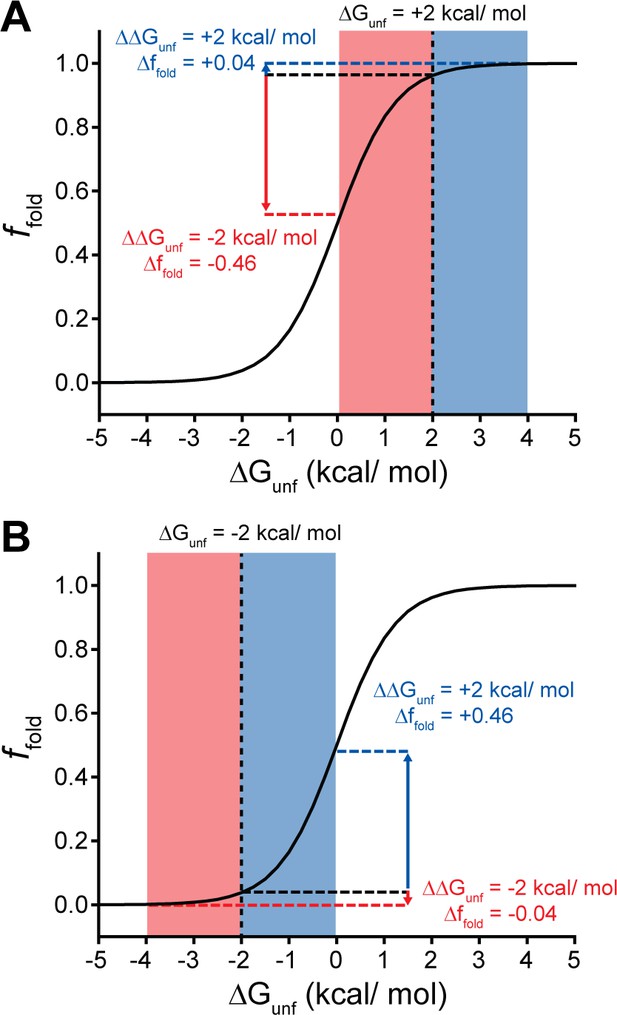
Context-dependent impacts of stabilizing and destabilizing mutations.
Line plots depict the relationship between the equilibrium fraction of folded protein (ffold) and the free energy of unfolding (ΔGunf). (A) A moderately stable protein (ΔGunf = +2 kcal/mol) exhibits a greater change in ffold after acquiring one or more destabilizing mutations (total ΔΔGunf = –2 kcal/mol, red) than after acquiring one or more mutations that stabilize the protein to the same extent (total ΔΔGunf = +2 kcal/mol, blue). (B) A moderately unstable protein (ΔGunf = –2 kcal/mol) exhibits a greater change in ffold after acquiring one or more stabilizing mutations (total ΔΔGunf = +2 kcal/mol, blue) than after acquiring one or more mutations that destabilize the protein to the same extent (total ΔΔGunf = –2 kcal/mol, red).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | GnRHR | GenBank | L01119 | - |
| Gene (Homo sapiens) | GnRHR | GenBank | L03380 | - |
| Strain, strain background (Escherichia coli) | NEB 10-β | New England Biolabs | C3020K | Electrocompetent |
| Cell line (H. sapiens) | HEK293T | ATCC | CRL-3216 | - |
| Transfected construct (H. sapiens) | Tet-Bxb1-BFP HEK293T | Laboratory of Doug Fowler | - | Clone 37 described in Jones et al., 2020 |
| Biological sample (Canis familiarus) | Pancreatic rough microsomes | tRNA probes | - | - |
| Biological sample (Oryctolagus cuniculus) | Reticulocyte lysate | Promega | L4960 | Nuclease-treated |
| Antibody | Anti-Hemagglutinin Antibody (mouse monoclonal) | Invitrogen | 26183-D550 | 2–2.2.14, DyLight 550 conjugate |
| Recombinant DNA reagent | pGEM- Lep- TMD6 | Hessa et al., 2007 | - | - |
| Recombinant DNA reagent | pcDNA5 CMV-HA-mGnRHR-IRES-eGFP | Chamness et al., 2021 | - | - |
| Recombinant DNA reagent | pcDNA5 attB-HA-mGnRHR-IRES-eGFP | This paper | - | A plasmid described in Chamness et al., 2021 was further modified as is described in ‘Materials and methods’ |
| Sequence-based reagent | Custom mGnRHR primer pool | Agilent Technologies Inc | - | See Supplementary file 3 for sequences |
| Sequence-based reagent | Custom 10- base randomization primer | Integrated DNA Technologies | - | gcatgaagaatctgcttagggttaggcgnnnnnnnnnncttcgcgatgtacgggccagat |
| Peptide, recombinant protein | PrimeSTAR HS DNA Polymerase | Takara Bio Inc | R010B | - |
| Peptide, recombinant protein | KAPA HiFi HotStart Ready Mix | Roche Diagnostics | 07958927001 | - |
| Commercial assay or kit | InFusion HD Cloning | Takara Bio Inc | 638944 | - |
| Commercial assay or kit | GenElute Mammalian gDNA Miniprep Kit | Sigma-Aldrich | G1N70 | - |
| Software, algorithm | Flowjo X | Treestar | - | - |
| Software, algorithm | OriginLab 2023 | OriginLab | - | - |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92406/elife-92406-mdarchecklist1-v1.pdf
-
Supplementary file 1
Predicted and measured apparent transfer free energies of mGnRHR TM6.
- https://cdn.elifesciences.org/articles/92406/elife-92406-supp1-v1.docx
-
Supplementary file 2
Solvent accessible surface area of mGnRHR residues.
- https://cdn.elifesciences.org/articles/92406/elife-92406-supp2-v1.docx
-
Supplementary file 3
mGnRHR library mutants with associated oligo sequences.
- https://cdn.elifesciences.org/articles/92406/elife-92406-supp3-v1.docx
-
Supplementary file 4
Deep mutational scanning measurements, epistasis scores, and cluster identities.
- https://cdn.elifesciences.org/articles/92406/elife-92406-supp4-v1.xlsx