Evolutionary adaptation to juvenile malnutrition impacts adult metabolism and impairs adult fitness in Drosophila
Figures
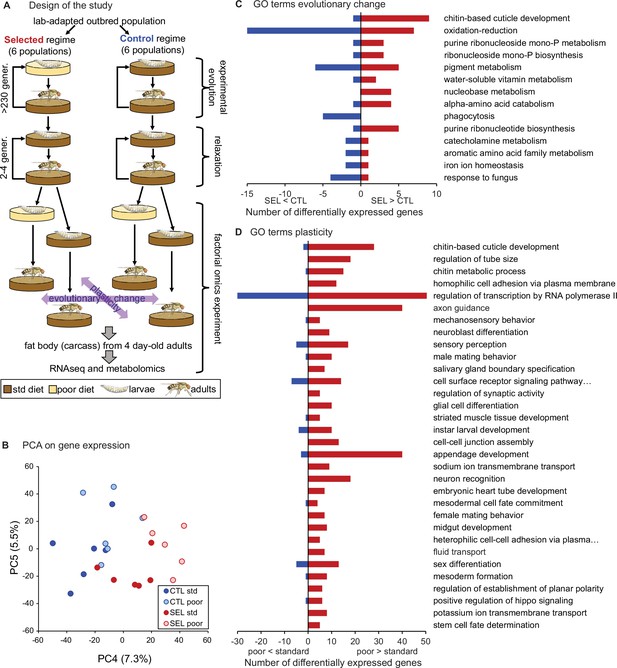
Phenotypic plasticity and genetically based evolutionary change in response to larval undernutrition both lead to divergence in gene expression patterns of adult Drosophila females.
(A) Design of the experimental evolution and of the gene expression assay. The purpose of ‘relaxation’ was to eliminate the potential effects of (grand)parental environment. The purple arrows indicate the two main factors in the analysis and interpretation, the evolutionary history (evolutionary change), and the effect of larval diet on which the current generation was raised (plasticity). (B) Sample score plot of the fourth and fifth principal components on the expression levels of 8701 genes of flies from Selected (SEL) and Control (CTL) populations raised on poor and standard (std) larval diet. Each point represents a replicate population × diet combination (Figure 1—source data 1). For complete plots of PC1-6, see Figure 1—figure supplement 1. (C) Biological process Gene Ontology (GO) terms enriched (at p < 0.01) in genes differentially expressed between Selected and Control populations, with the number of genes upregulated (red) and downregulated (blue) in Selected populations. (D) GO terms enriched at p < 0.01 among genes showing a plastic response to diet. For details of the GO term enrichment results, see Supplementary file 2.
-
Figure 1—source data 1
Principal component analysis of adult gene expression: the first six principal component scores of all populations on each diet.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig1-data1-v2.xlsx
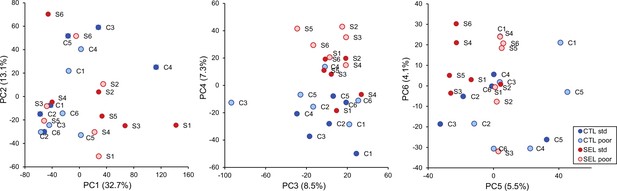
Plots of the first six principal components of the gene expression levels of adults from the Selected and Control populations raised on the standard and poor diet (Figure 1—source data 1).
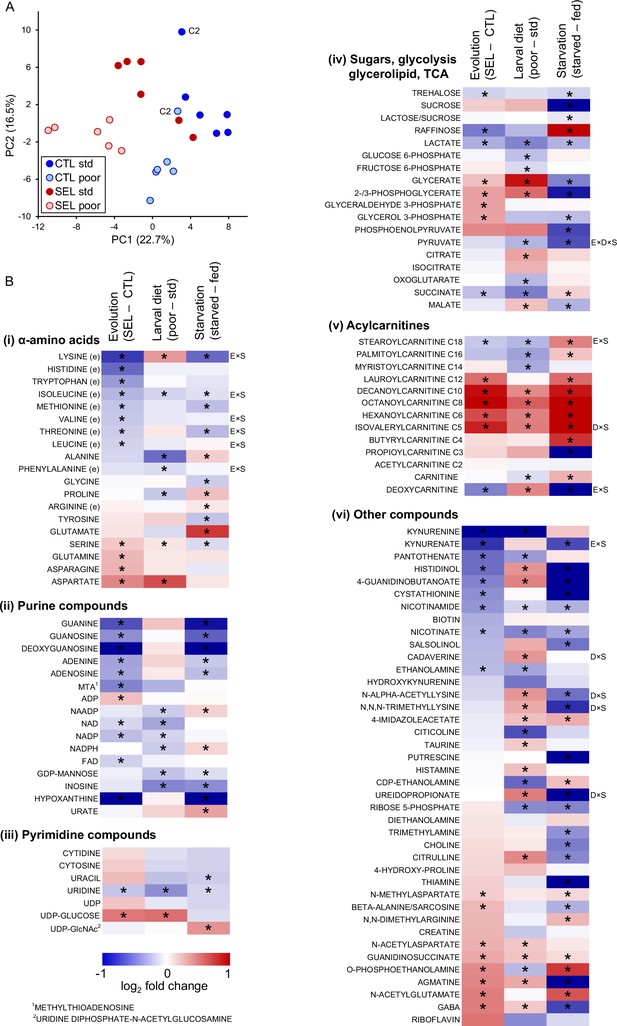
The effects of phenotypic plasticity and evolutionary change on adult metabolite abundance.
(A) Principal component score plot based on metabolic signatures composed of 113 robustly quantified metabolites in flies from the six Selected and six Control populations raised on both poor and standard diet (Figure 2—source data 1). The flies were in fed condition. Metabolite abundances for two replicate samples were averaged before the principal component analysis (PCA); thus, there is one point per population and diet. The ‘C2’ label indicates control population 2 on either diet; this population deviated from the other Control populations along the PC2 axis; however, we retained it for the univariate analysis. (B) The effects of experimental factors on abundance of individual metabolites. The first two columns of the heat maps indicate, respectively, the effect of evolutionary regime (least square mean difference Selected – Control) and the within-generation (phenotypically plastic) effect of larval diet (least square mean difference poor – standard diet) on the relative concentration of metabolites in fed flies. The third column indicates the main effect of the starvation treatment (least square mean difference between starved and fed flies) on metabolite concentrations in flies from both Selection regimes raised on both diets. Thus, red (blue) indicates that a compound is more (less) abundant in Selected, poor diet-raised and starved flies than in Control, standard diet-raised and fed flies, respectively. Asterisks indicate effects significant at q < 0.1. Annotations to the right of the heat maps indicate interactions significant at q < 0.1: E × S = evolutionary regime × starvation treatment, D × S = larval diet × starvation treatment, E × D × S = three-way interaction; (e) indicates an essential amino acid, C2–C18 for acylcarnitines refers to the length of the acyl chain. For estimates, effects, and statistical tests underlying the figure, see Supplementary file 3; original data are in Supplementary file 8.
-
Figure 2—source data 1
Principal component analysis of adult metabolite abundance in fed flies: the first four principal component scores of all populations.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig2-data1-v2.xlsx
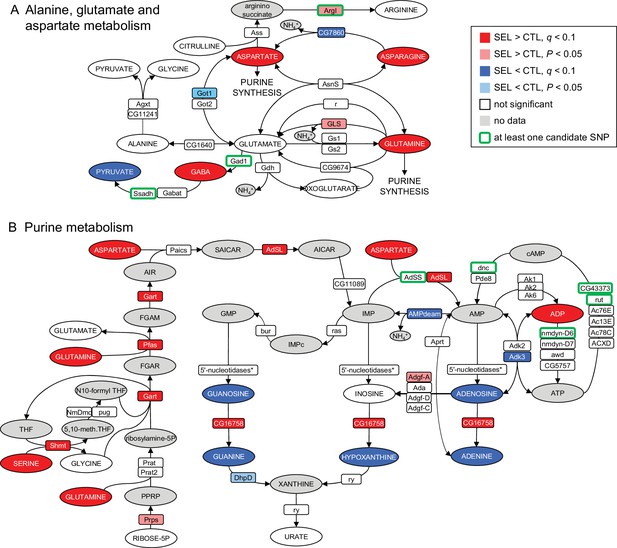
Imprint of evolutionary adaptation to poor larval diet on (A) alanine, glutamate, and aspartate metabolism and (B) purine metabolism pathways of adult female flies.
Highlighted in color are metabolites differentially abundant and enzymes differentially expressed between the Selected and Control populations, as well as enzymes to which at least one single-nucleotide polymorphisms (SNP) differentiated in frequency between the sets of populations has been annotated. Enzymes differentially expressed at the nominal p < 0.05 but not passing the 10% false discovery rate (FDR) are also indicated. Metabolites in gray were either not targeted, not detected, or the data did not pass quality filtering. The pathways have been downloaded from KEGG (https://www.genome.jp/kegg/pathway.html). For the sake of clarity, some substrates and products of depicted reactions that were not differentially abundant or were absent from the data, as well as cofactors, have been omitted. Only genes expressed in female fat body have been included. *5'-nucleosidases: cN-IIB, NT5E-2, Nt5B, veil, CG11883; none was differentially expressed.
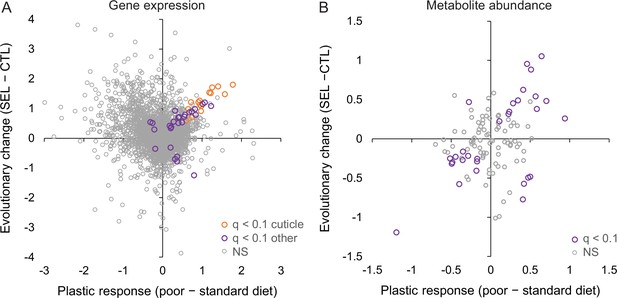
Relationship between phenotypic plasticity and genetically based evolutionary change of adult gene expression (A) and metabolite abundance (B) in response to larval diet.
The plastic responses and evolutionary change for each gene and metabolite were estimated, respectively, as the main effect of larval diet and evolutionary regime in the factorial statistical models (see ‘Materials and methods’). Color highlights genes and metabolites with significant effects (q < 0.1) for both diet and regime; in (A), orange symbols indicate significant genes involved in cuticle, integument, or tracheal development (Figure 4—source data 1).
-
Figure 4—source data 1
The estimated main effects of diet (the plastic response) and of evolutionary regime (the evolutionary change) on adult gene expression and metabolite abundance.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig4-data1-v2.xlsx
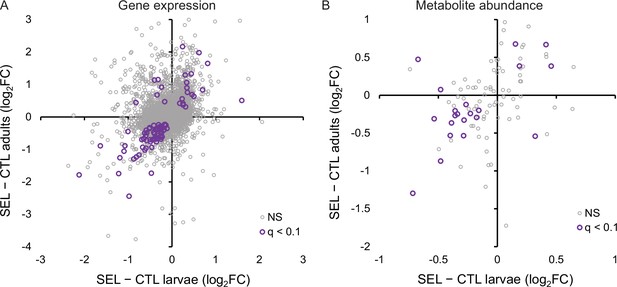
The relationship between the evolutionary change (i.e., the difference between the Selected and Control populations) in gene expression and metabolite abundance at the larval and adult stage.
(A) Correlation of evolutionary changes in gene expression (all genes: r = 0.35, N = 8437, p < 0.0001; genes with q < 0.1: r = 0.74, N = 84, p < 0.0001). Five genes (not passing q < 0.1) are outside of the range of the plot. (B) Correlation of evolutionary changes in metabolite abundance (all metabolites: r = 0.37, N = 97, p = 0.0002; metabolites with q < 0.1: r = 0.55, N = 21, p = 0.0091). The estimates plotted are for larvae and adults raised on the poor larval diet (see ‘Materials and methods’); q < 0.1 refers to genes or metabolites that pass the 10% false discovery rate (FDR) threshold for both stages; NS are the remaining genes or metabolites (Figure 5—source data 1, sheet A and B).
-
Figure 5—source data 1
The relationship between evolutionary change in adults and the corresponding change in the larvae.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig5-data1-v2.xlsx
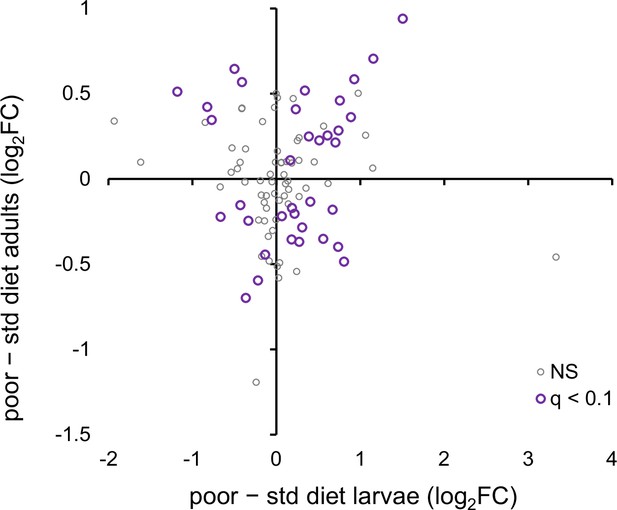
The relationship between the phenotypically plastic responses of the metabolomes of larvae and adults to larval diet.
The plotted values are main effect estimates from the statistical models (i.e., they are averaged across the Selected and Control populations). q < 0.1 refers to metabolites that pass the 10% false discovery rate (FDR) threshold for both stages; NS are the remaining metabolites (Figure 5—source data 1, sheet C).
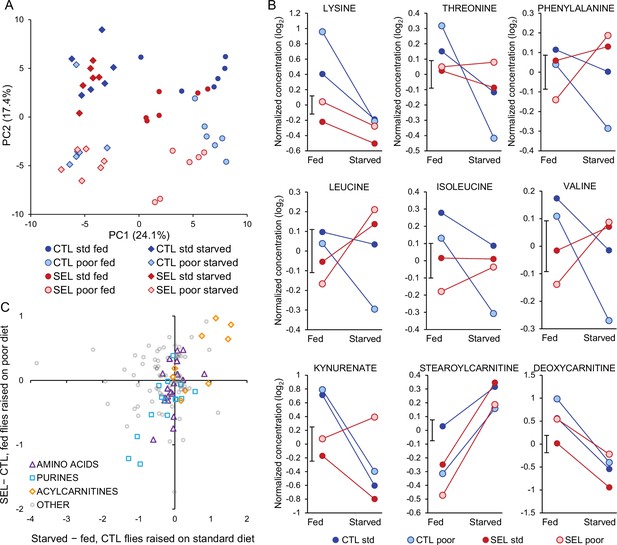
Effects of starvation on metabolite abundance.
(A) Principal component score plot on metabolite abundance for all conditions (Selected versus Control, poor versus standard larval diet, fed versus starved flies). The two replicate samples from the same population × diet × starvation treatment combination were averaged (Figure 6—source data 1). (B) Metabolites that show significant (q < 0.1) interaction between the evolutionary regime and starvation treatment. Plotted are least-square means obtained from the full factorial general mixed model (Figure 6—source data 2). The bar next to the Y-axis indicates ± standard error of the least-square means (i.e., its width corresponds to 2 SEM); the standard error is identical for all treatments because the least-square means were estimated from the general mixed model and the design is balanced. For compounds showing other interactions, see Figure 6—figure supplement 1. (C) The relationship between the effect of 24 hr starvation (starved minus fed flies) on metabolite abundance in Control flies raised on the standard diet and the evolutionary change (Selected minus Control) quantified in fed flies raised on poor diet (Figure 6—source data 3). Three categories of metabolites of interest are highlighted; ‘acylcarnitines’ only include those with even-numbered acyl chain (resulting from catabolism of triglycerides) but not those with 3C and 5C chains (which are products of amino acid catabolism). Pearson’s correlations: amino acids r = 0.50, N = 19, p = 0.030; purine compounds r = 0.76, N = 16, p = 0.0006; acylcarnitines r = 0.74, N = 9, p = 0.022; other metabolites r = 0.02.
-
Figure 6—source data 1
Principal component analysis of metabolite abundance in fed and starved flies: the first four principal component scores of all populations under all conditions.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Abundance patterns of metabolites with interactions.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Relationship between response to starvation and the evolutionary change.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig6-data3-v2.xlsx
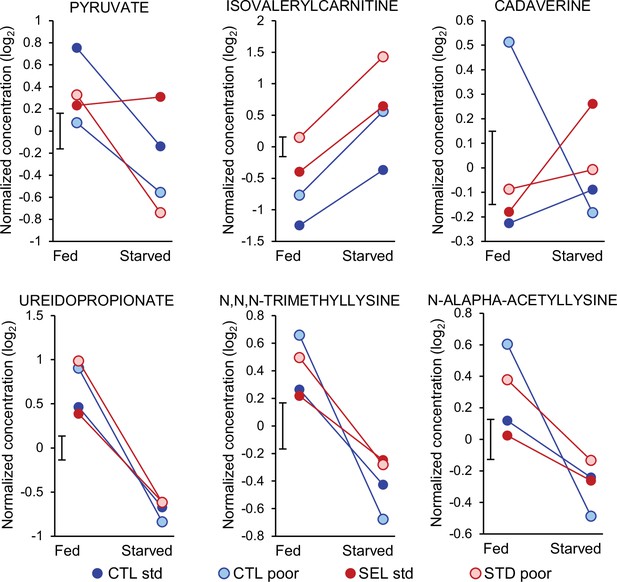
Metabolites showing a significant three-way interaction evolutionary regime × larval diet × starvation (pyruvate) or the two-way interaction between larval diet and starvation treatment (the remaining five compounds).
q < 0.1 was used as the significance threshold. Symbols indicate least-square means obtained from the full factorial general mixed model (Figure 6—source data 2). The bar next to the Y-axis indicates the standard error of the means (identical for all treatments as they were estimated from the model).
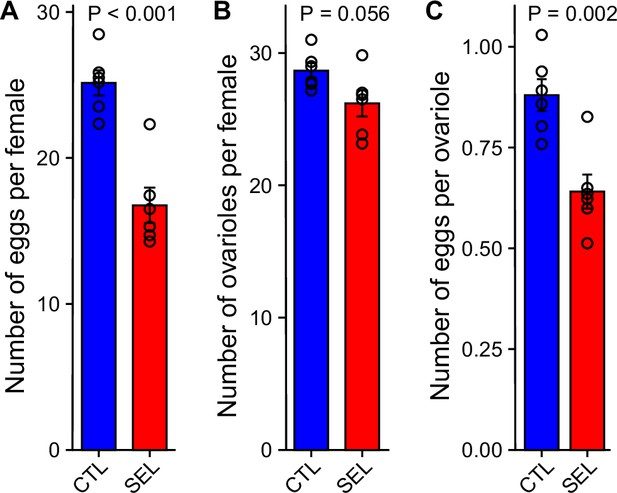
Selected populations pay a fecundity cost for larval adaptation to poor diet.
(A) The number of eggs laid overnight by females from the Selected and Control (GMM; F1,8 = 44.5, p = 0.0002, N = 16 per evolutionary regime, 2–3 per replicate population). (B) The number of ovarioles (left + right ovary) per female (GMM; F1,10 = 4.7, p = 0.056, N = 72 per regime, six per replicate population). (C) The mean number of eggs per ovariole (one-way ANOVA; F1,10 = 17.1, p = 0.002, N = 6 populations per regime). The females experienced the dietary conditions, under which the Selected populations evolved: the larvae were raised on the poor diet; the adults were transferred to the standard diet and additionally provided live yeast 24 hr before oviposition. Bars are means, error bars correspond to ± SEM, the symbols to means of replicate populations (Figure 7—source data 1).
-
Figure 7—source data 1
Data on fecundity and ovariole number.
- https://cdn.elifesciences.org/articles/92465/elife-92465-fig7-data1-v2.xlsx
Additional files
-
Supplementary file 1
Results of univariate analysis of gene expression in adult female carcass performed in limma-voom.
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp1-v2.xlsx
-
Supplementary file 2
Results of GO term enrichment for ‘biological process’ on genes expressed in female adult carcass.
(A) Genes differentially expressed between Selected and Control flies. (B) Genes differentially expressed in flies raised on standard versus poor larval diet. (C) Genes differentially expressed between Selected and Control populations both in adults and larvae. (D) Genes differentially expressed between Selected and Control populations in adults but not in larvae.
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp2-v2.xlsx
-
Supplementary file 3
Results of univariate analysis of metabolome with general mixed model.
(A) Least-square means for all combinations of the three experimental factors: evolutionary regime (Control versus Selected), larval diet (standard versus poor), and adult starvation treatment (fed versus starved). The LS means and standard errors (SE) were obtained from the full GMM model. They are expressed on a zero-centered log2 scale. (B) Least-square means contrasts corresponding to Figure 2B: Regime(Fed) = Selected – Control in fed condition; Diet(Fed) = flies in fed condition raised on standard diet – flies in fed condition raised on poor diet; Starv = starved flies – fed flies across both regimes and diets. (C) Tests of significance (df, F, nominal P, q = adjusted P) from separate analysis on flies in fed condition only. (D) Tests of significance from the full model on both fed and starved flies. Denominator degrees of freedom (df) were estimated with the Satterthwaite method and thus vary across compounds depending on the magnitude of variance components corresponding to random factors.
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp3-v2.xlsx
-
Supplementary file 4
Results of joint pathway analysis on metabolites significantly different between Selected and Control flies (in fed state).
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp4-v2.xlsx
-
Supplementary file 5
Genes that show differential expression between Selected and Control populations (at raw p < 0.05) at both larval and adult stage.
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp5-v2.xlsx
-
Supplementary file 6
Analysis of the effect of evolutionary regime on the expression of genes involved in amino acid metabolism and purine synthesis in the larvae.
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp6-v2.xlsx
-
Supplementary file 7
MANOVA on the first two principal component scores from the principal component analysis on metabolite abundance in fed and starved flies.
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp7-v2.xlsx
-
Supplementary file 8
Original data on metabolite abundance.
The table includes the original estimates of peak area (in arbitrary units) normalized to the sample protein content, and log10-transformed, zero-centered, and Pareto-scaled values used for the analysis (zero-centering and Pareto-scaling was done separately for each batch). The table includes the 13 outlier data points that were removed from the final analysis because their Studentized residuals exceeded ±4; these outliers are identified in the last column.
- https://cdn.elifesciences.org/articles/92465/elife-92465-supp8-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92465/elife-92465-mdarchecklist1-v2.docx





