Developmental conversion of thymocyte-attracting cells into self-antigen-displaying cells in embryonic thymus medulla epithelium
Figures
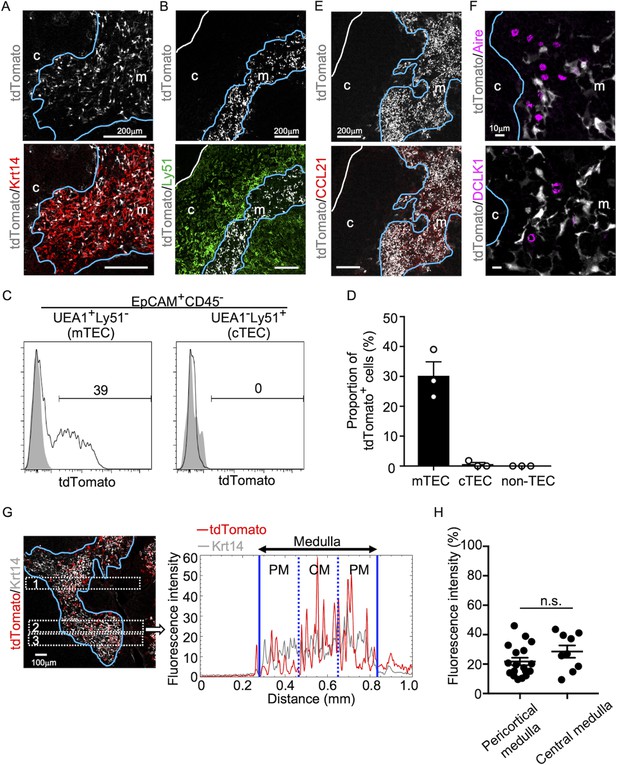
CCL21-expressing medullary thymic epithelial cells (mTECs) in postnatal thymus.
Immunofluorescence analysis of thymus sections from 5-week-old Ccl21atdTomato/+ mice. tdTomato fluorescence (white) was detected with Keratin 14 (Krt 14, red) (A) or Ly51 (green) (B). White lines indicate capsular outline of the thymus. Blue lines show cortico-medullary junction. Representative data from at least three independent experiments are shown. c, cortex. m, medulla. Scale bar, 200 μm. (C) Flow cytometric profiles of tdTomato fluorescence expression in EpCAM+CD45−UEA1+Ly51− mTECs (left) and EpCAM+CD45−UEA1−Ly51+ cTECs (cortical thymic epithelial cells) (right) from Ccl21atdTomato/+ mice (black line histograms) and WT mice (gray shaded histograms). Numbers in histograms indicate frequency of cells within indicated area. (D) Proportions (means and standard error of the means [SEMs], n = 3) of tdTomato+ cells in EpCAM+CD45−UEA1+Ly51− mTECs, EpCAM+CD45−UEA1−Ly51+ cTECs, and EpCAM− non-TECs analyzed as in (C). (E, F) Immunofluorescence analysis of thymus sections from 5-week-old Ccl21atdTomato/+ mice. tdTomato fluorescence (white) was detected with CCL21 (red) (E), Aire, or DCLK1 (magenta) (F). White lines indicate capsular outline of the thymus. Blue lines show cortico-medullary junction. Representative data from at least three independent experiments are shown. c, cortex. m, medulla. Scale bar, 200 μm (E) or 10 μm (F). (G) Distribution of tdTomato-expressing cells in the medullary region defined by Krt14 expression. Left panel shows representative tdTomato (red) and Krt14 (white) fluorescence signals in the thymus sections from Ccl21atdTomato/+ mice. Scale bar, 100 μm. Right panel shows fluorescence intensity profiles of tdTomato (red line) and Krt14 (gray line) signals within the region of interest (ROI) defined by dashed rectangles in the left panel. Medullary regions were equally divided into three areas into pericortical areas (PM) and central area (CM). (H) tdTomato intensity (means and standard error of the means [SEMs], n = 3) in indicated areas (G) and Figure 1—figure supplement 1A were calculated in comparison with total tdTomato intensity within the ROI. n.s., not significant.

Distribution of Ccl21a-expressing cells in postnatal thymus.
A replicate of thymus section analysis for postnatal Ccl21atdTomato/+ mice as in Figure 1E.
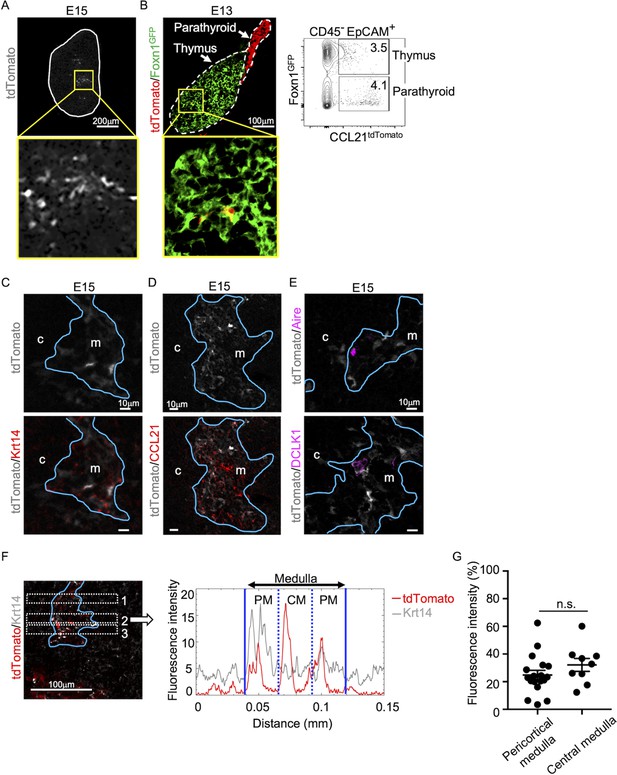
CCL21-expressing medullary thymic epithelial cells (mTECs) in embryonic thymus.
(A) tdTomato fluorescence (white) detected in thymus sections from E15 Ccl21atdTomato/+ embryos. White lines indicate the capsular outline of the thymus. The image in central region identified by the box in upper panel is magnified in bottom panel. Representative data from at least three independent experiments are shown. Scale bar, 200 μm. (B) tdTomato (red) and GFP (green fluorescence protein) (green) fluorescence in thymus sections from E13 Ccl21atdTomato/+ × Foxn1-GFP-transgenic embryos (left). Dashed lines show the outlines for the thymus and parathyroid primordia. Central region of the thymus identified by the box in upper panel is magnified in bottom panel. Scale bar, 100 μm. Right panel shows a representative flow cytometric profile for tdTomato and GFP in CD45−EpCAM+ epithelial cells. Numbers in the contour plot indicate frequency of cells within indicated area. (C–E) Immunofluorescence analysis of thymus sections from E15 Ccl21atdTomato/+ embryos. tdTomato fluorescence (white) was detected with Krt14 (red) (C), CCL21 (red) (D), Aire, or DCLK1 (magenta) (E). Blue lines show the cortico-medullary junction. Representative data from at least three independent experiments are shown. c, cortex. m, medulla. Scale bar, 10 μm. (F) Distribution of tdTomato-expressing cells in the medullary region defined by Krt14 expression. Left panel shows representative tdTomato (red) and Krt14 (white) fluorescence signals in the thymus sections from Ccl21atdTomato/+ E15 embryos. Scale bar, 100 μm. Right panel shows fluorescence intensity profiles of tdTomato (red line) and Krt14 (gray line) signals within the region of interest (ROI) defined by dashed rectangles in the left panel. The medullary regions were equally divided into three areas into pericortical areas (PM) and central area (CM). (G) tdTomato intensity (means and standard error of the means [SEMs], n = 3) in indicated areas (F) and Figure 1B were calculated in comparison with total tdTomato intensity within the ROI. n.s., not significant.
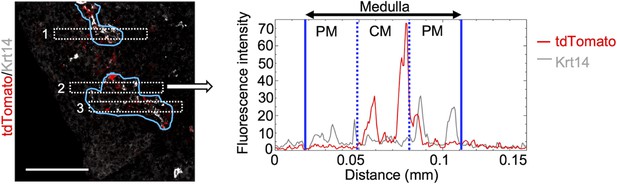
Distribution of Ccl21a-expressing cells in embryonic thymus.
A replicate of thymus section analysis for E15 Ccl21atdTomato/+ mice as in Figure 2F.
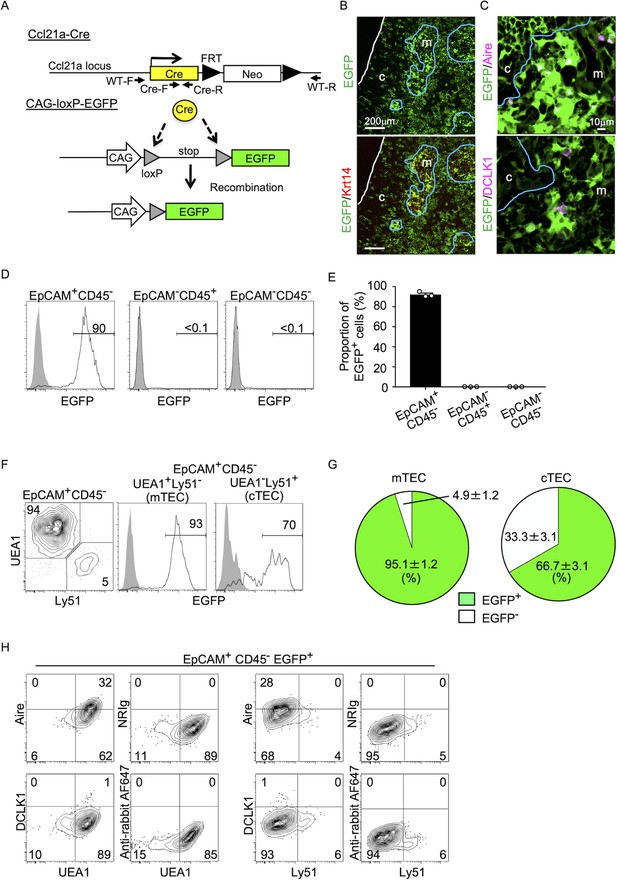
Fate mapping of Ccl21a-expressing cells in the thymus.
(A) Schematic illustration of Cre-dependent Ccl21a-specific EGFP expression in Ccl21a-Cre × CAG-loxP-stop-loxP-EGFP mice. FRT, flippase recognition target site. Neo, neomycin resistance gene. Arrows indicate PCR primers for allele genotyping. (B, C) Immunofluorescence analysis of thymus sections from 3-week-old Ccl21a-Cre × CAG-loxP-EGFP mice. EGFP fluorescence (green) was detected with Krt14 (red) (B), Aire, or DCLK1 (magenta) (C). White lines indicate the capsular outline of the thymus. Blue lines show the cortico-medullary junction. Representative data from at least three independent experiments are shown. c, cortex. m, medulla. Scale bar, 200 μm (B) or 10 μm (C). (D) Flow cytometric analysis of liberase-digested thymus cells isolated from 3-week-old Ccl21a-Cre × CAG-loxP-EGFP mice. Histograms show EGFP fluorescence expression in EpCAM+CD45− cells (left), EpCAM−CD45+ cells (middle), and EpCAM−CD45− cells (right) prepared from Ccl21a-Cre × CAG-loxP-EGFP mice (black histograms) and littermate control (gray shaded histograms). Numbers in histograms indicate frequency of cells within indicated area. (E) Proportions (means and standard error of the means [SEMs], n = 3) of EGFP+ cells in indicated cell populations analyzed as in (D). (F) Flow cytometric profiles of UEA1 reactivity and Ly51 expression in EpCAM+CD45− TECs (left). Histograms show EGFP fluorescence expression in EpCAM+CD45−UEA1+Ly51− medullary thymic epithelial cells (mTECs; middle) and EpCAM+CD45−UEA1−Ly51+ cTECs (right) from Ccl21a-Cre × CAG-loxP-EGFP mice (black histograms) and littermate control (gray shaded histograms). Numbers in the histograms indicate frequency of cells within indicated area. (G) Proportions (means and SEMs, n = 3) of EGFP+ and EGFP− cells in EpCAM+CD45−UEA1+Ly51− mTECs (left) and EpCAM+CD45−UEA1−Ly51+ cTECs (right) analyzed as in (F). (H) Flow cytometric analysis of liberase-digested thymus cells isolated from Ccl21a-Cre × CAG-loxP-EGFP mice at 3 weeks old. Contour plots show Aire (top), DCLK1 (bottom), or isotype control detection with UEA1 reactivity or Ly51 expression in EpCAM+CD45−EGFP+ TECs. Numbers in the plots indicate frequency of cells within the indicated area. Representative data from three independent experiments are shown.
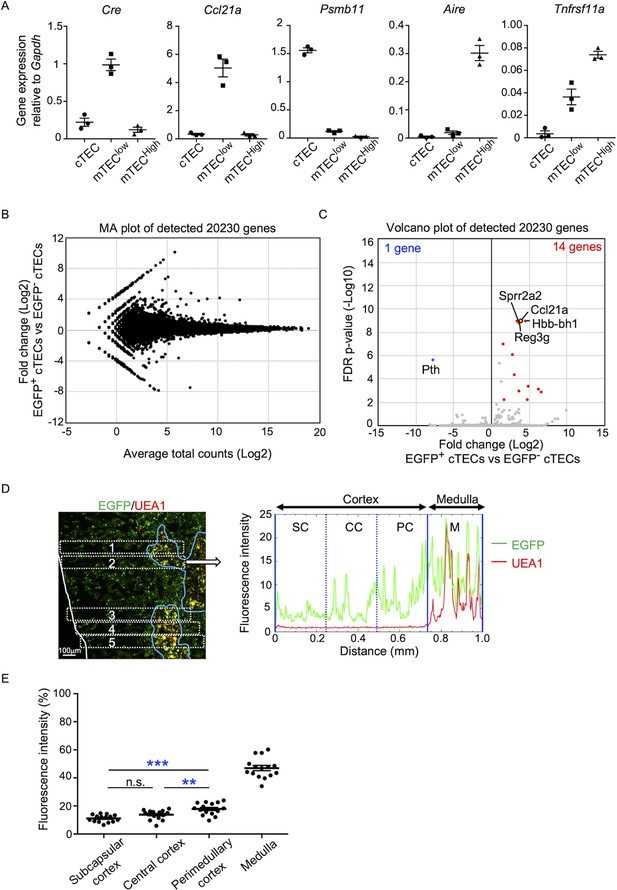
Characterization of cTECs that previously transcribed Ccl21a.
(A) Quantitative RT-PCR analysis of Cre and indicated genes (means and standard error of the means [SEMs], n = 3) in cTECs (EpCAM+CD45−UEA1−Ly51+), mTEClow (EpCAM+CD45−UEA1+Ly51− I-Alow), and mTEChigh (EpCAM+CD45−UEA1+Ly51− I-Ahigh) isolated from Ccl21aCre/+ mice. (B) MA plot of 20,230 genes detected in RNA-sequencing analysis of EGFP+ and EGFP− cTECs isolated from Ccl21a-Cre × CAG-loxP-EGFP mice at 2-week-old. Detected genes are plotted as log2 average total counts versus log2 fold changes (EGFP+ cTECs/EGFP− cTECs). (C) Volcano plot analysis of EGFP+ and EGFP− cTECs. Detected genes are plotted as log2 fold changes (EGFP+ cTECs/EGFP− cTECs) versus −log10 FDR p-values. Fourteen genes (red symbols) are more highly detected (log2 fold change >1.7, FDR p-value <0.05) in EGFP+ cTECs than EGFP− cTECs, whereas one gene (blue symbol) is more highly detected (log2 fold change <−1.7, FDR p-value <0.05) in EGFP− cTECs than EGFP+ cTECs. (D) Distribution of EGFP-expressing cells in the thymus. Left panel shows representative fluorescence signals for EGFP (green) in the thymus sections from Ccl21a-Cre × CAG-loxP-EGFP mice. The medullary region is defined by UEA1 reactivity (red). White lines indicate the capsular outline of the thymus. Blue lines show the cortico-medullary junction. Scale bar, 100 μm. Right panel shows fluorescence intensity profiles of EGFP (green line) and UEA1 (gray line) signals within the region of interest (ROI) defined by dashed rectangles in the left panel. Cortical regions were equally divided into three areas into subcapsular cortex (SC), central cortex (CC), and perimedullary cortex (PC). M, medulla. (E) EGFP intensity (means and SEMs, n = 3) in indicated areas (C) and Figure 1C were calculated in comparison with total EGFP intensity within the ROI. n.s., not significant. **p < 0.01, ***p < 0.001.

Purity of isolated EGFP+ and EGFP− cTECs for transcriptomic analyses.
Flow cytometric analysis of indicated cells (n = 3) from Ccl21a-Cre × CAG-loxP-EGFP mice at 2-week-old. Numbers indicate frequency of cells within indicated areas.
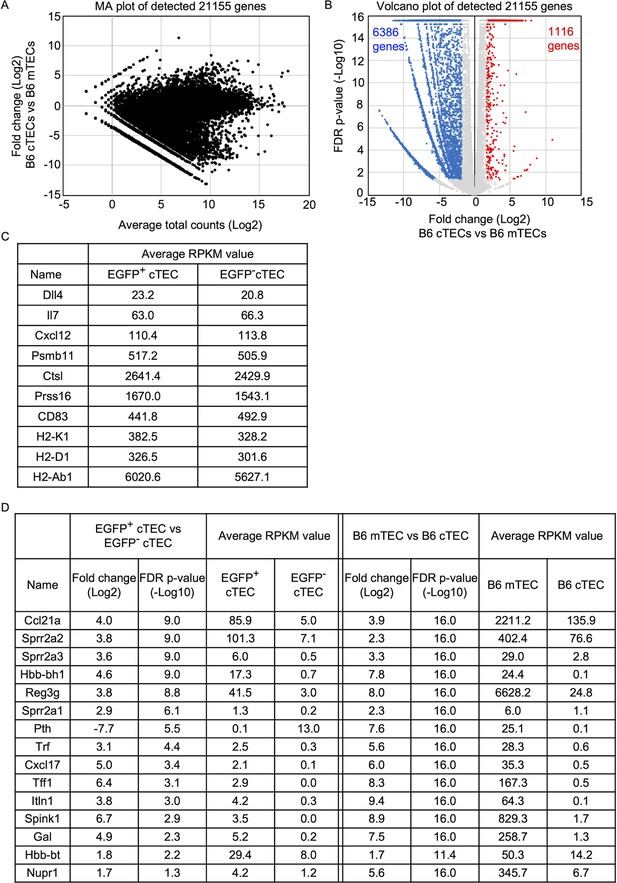
Transcriptomic profiles of cTECs, medullary thymic epithelial cells (mTECs), EGFP+ cTECs, and EGFP− cTECs.
(A) MA plot of 21,155 genes detected in RNA-sequencing analysis of cTECs and mTECs isolated from B6 mice (Ohigashi et al., 2019). Detected genes are plotted as log2 average total counts versus log2 fold changes (cTECs/mTECs). (B) Volcano plot analysis of cTECs and mTECs. Detected genes are plotted as log2 fold changes (EGFP+ cTECs/EGFP− cTECs) versus −log10 FDR p-values. 1116 genes (red symbols) are more highly detected (log2 fold change >1.7, FDR p-value <0.05) in cTECs than mTECs, whereas 6386 gene (blue symbol) is more highly detected (log2 fold change <−1.7, FDR p-value <0.05) in mTECs than cTECs. (C) Average RPKM value of indicated genes detected in EGFP+ and EGFP− cTECs from Ccl21a-Cre × CAG-loxP-EGFP mice. (D) Log2 fold changes, −log10 FDR p-values, and average RPKM values of genes differently detected between EGFP+ and EGFP− cTECs from Ccl21a-Cre × CAG-loxP-EGFP mice, and those values in RNA-sequencing analysis of mTECs and cTECs from B6 mice.

Distribution of EGFP+ cells in the thymus.
(A) A replicate of thymus section analysis for postnatal Ccl21a-Cre × CAG-loxP- EGFP mice as in Figure 4D. (B) Distribution of Ly51 in the thymus. Left panel shows representative fluorescence signals for Ly51 (red) and EGFP (green) in the thymus sections from Ccl21a-Cre × CAG-loxP-EGFP mice. Right panel shows fluorescence intensity profiles of Ly51 (red), EGFP (green), and UEA1 (blue) signals within the region of interest (ROI) defined by dashed rectangles in left panel. Cortical regions were equally divided into three areas as in Figure 4D. (C) Ly51 signal intensity (means and standard error of the means [SEMs], n = 3) in indicated areas (D) were calculated in comparison with total Ly51 signal intensity within the ROI. n.s., not significant.
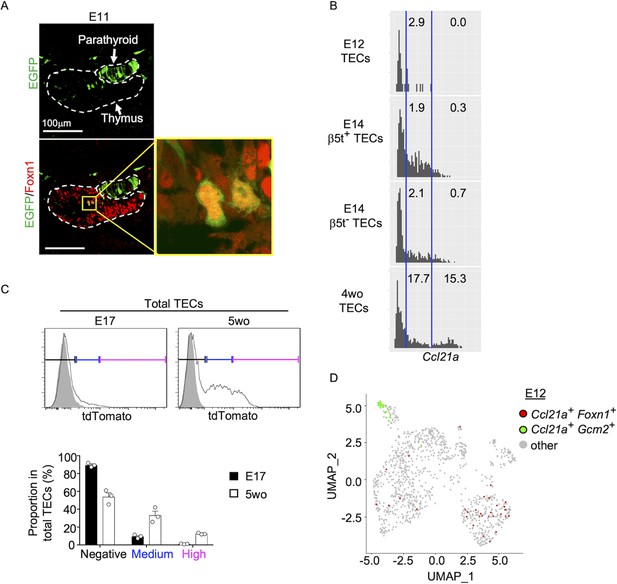
Ccl21a-expressing medullary thymic epithelial cells (mTECs) during early thymus organogenesis.
(A) Immunofluorescence analysis of thymus section from Ccl21a-Cre × CAG-loxP-EGFP E11 embryos. EGFP (green) and Foxn1 (red) were analyzed as indicated. Dashed lines show the outline of the thymus and parathyroid primordia. Image in yellow box in the middle of the thymus is magnified in right panel. Scale bar, 100 μm. Representative data from two independent experiments are shown. (B) Single-cell RNA-sequencing analysis of Epcam+Foxn1+ TECs from E12, E14, and 4-week-old β5t-Venus knock-in mice. Normalized log transcript counts of Ccl21a mRNA (x-axis) and cell numbers (y-axis) are plotted. For E14 TECs, β5t-Venus+ and β5t-Venus− TECs were sorted and analyzed in parallel. Numbers in histograms indicate frequency of cells within indicated area. (C) Flow cytometric analysis of tdTomato expression in EpCAM+CD45− TECs isolated from E17 embryonic and 2-week-old postnatal Ccl21atdTomato/+ mice (black histograms) and WT mice (gray shaded histograms). Bottom plots indicate the proportions (means and standard error of the means [SEMs], n = 3) of tdTomatonegative (black), tdTomatomedium (blue), and tdTomatohigh (magenta) cells within EpCAM+CD45− TECs as defined in top histograms. (D) Single-cell RNA-sequencing analysis of Epcam+ E12 embryonic pharyngeal epithelial cells. Dots indicate Uniform Manifold Approximation and Projection (UMAP) plot of Ccl21a+Foxn1+ mTECs (red), Ccl21a+Gcm2+ parathyroid epithelial cells (green), and other cells (gray).
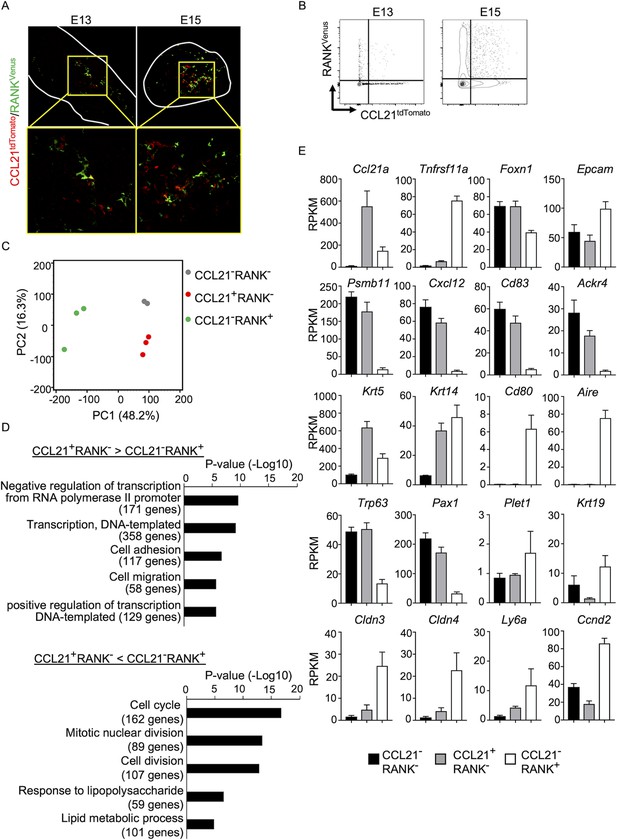
CCL21-expressing medullary thymic epithelial cells (mTECs) are distinct from RANK-expressing mTECs.
(A) Immunofluorescence analysis of tdTomato (red) and Venus (green) signals in thymus sections from Ccl21atdTomato/+Tnfrsf11a(RANK)Venus mice at indicated embryonic age. White lines indicate the capsular outline of the thymus. Images in boxed regions in upper panels are magnified in bottom panels. Representative data from at least three independent experiments are shown. (B) Flow cytometric profiles of tdTomato and Venus fluorescence signals in EpCAM+CD45− TECs isolated from Ccl21atdTomato Tnfrsf11a(RANK)Venus mice at indicated embryonic age. (C) Principal component analysis of RNA-sequencing data of indicated cell populations isolated from Ccl21atdTomato/+Tnfrsf11a(RANK)Venus E17 embryos. (D) Enrichment analysis of ontology for genes that are differently expressed (RPKM >1, fold change >1.5 or <—1.5, FDR p-value <0.05) between CCL21+RANK− and CCL21−RANK+ TECs. Bars show the adjusted p-values of the top five categories enriched in CCL21+RANK− TECs (top) and CCL21−RANK+ TECs (bottom). Numbers in parentheses indicate the number of categorized genes. (E) RPKM values of indicated genes detected in RNA-sequencing analysis.
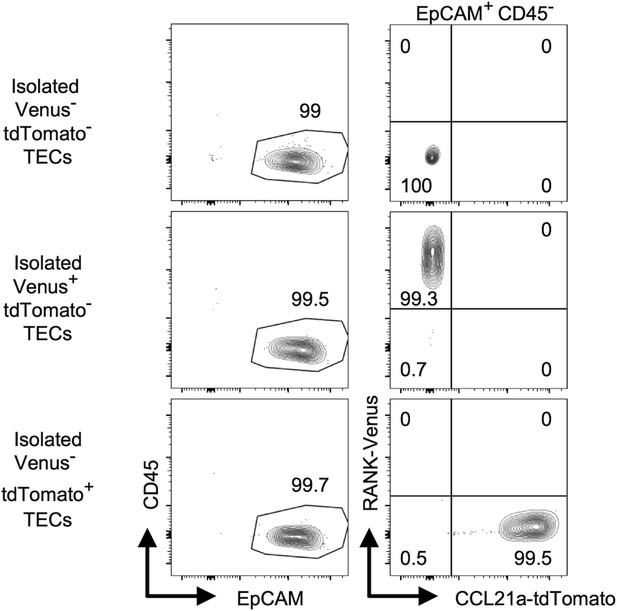
Purity of isolated TECs for transcriptomic analysis.
Shown are flow cytometric profiles of indicated cells from Ccl21atdTomato Tnfrsf11a(RANK)Venus E17 mice. Numbers indicate frequency of cells within indicated areas.
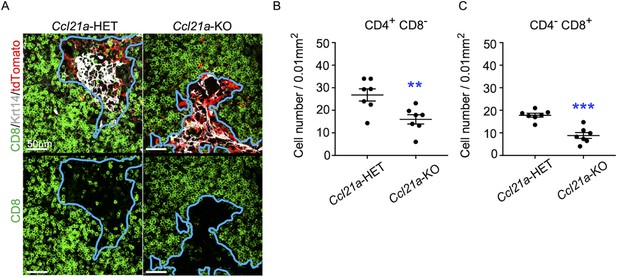
CCL21-expressing medullary thymic epithelial cells (mTECs) in E17 embryos are functional in thymocyte attraction.
(A) Immunofluorescence analysis of CD4, CD8, Krt14, and tdTomato in thymus sections from heterozygous (Ccl21atdTomato/+) control mice or homozygous (Ccl21atdTomato/tdTomato) CCL21-deficient mice at E17. E17 thymus sections were stained for CD4, CD8, and Krt14. Representative images for Krt14 (white), tdTomato fluorescence (red), and CD8 (green) expressions are shown. Blue lines indicate cortico-medullary junctions, according to the distribution of Krt14-expressing mTECs. tdTomato-positive cells were localized in the medullary regions. Scale bar, 50 μm. (B, C) Graphs show the means and standard error of the means (SEMs; n = 3) of the numbers of CD4+CD8− (B) and CD4−CD8+ (C) thymocytes per unit area (0.01 mm2) in the medullary regions. **p < 0.01; ***p < 0.001.
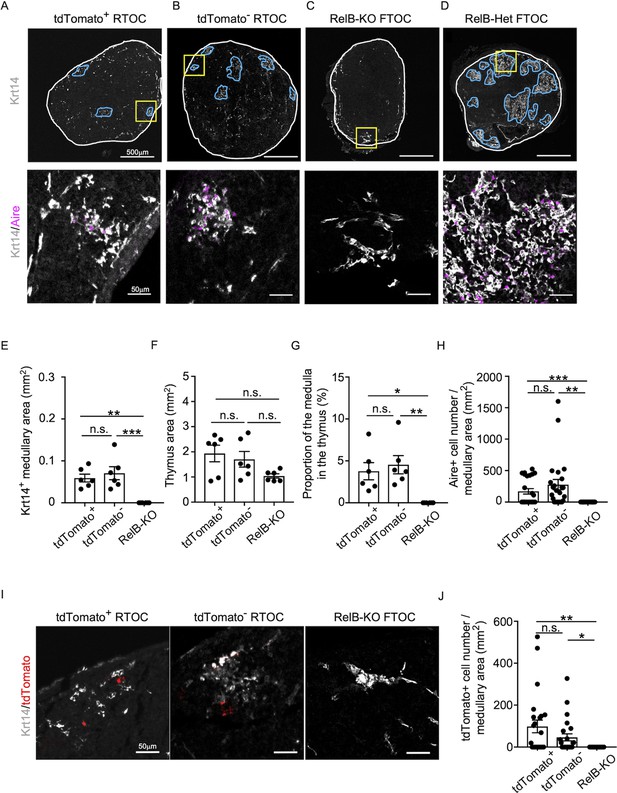
Developmental potential of CCL21-expressing medullary thymic epithelial cells (mTECs).
(A–D) Immunofluorescent staining of indicated thymus graft sections. Shown in upper panels are Krt14 expression (white). White lines indicate capsular outline of the thymus. Blue lines show Krt14+ medullary region. Scale bar, 500 μm. Bottom panels show Krt14 (white) and Aire (magenta) fluorescence signals within yellow boxes in upper panels. Scale bar, 50 μm. Grafts represent RelB-KO thymic stroma reaggregated with tdTomato+ TECs (A) and tdTomato− TECs (B) as well as fetal thymus lobe from RelB-KO mice (C) and RelB-heterozygous mice (D). (E) Size of Krt14+ medullary areas (means and standard error of the means [SEMs], n = 6) in indicated thymus graft sections. (F) Size of grafted thymus areas (means and SEMs, n = 6) in indicated thymus graft sections. (G) Proportion (means and SEMs, n = 6) of Krt14+ medullary areas in the thymus areas in indicated thymus graft sections. (H) Numbers of Aire+ mTECs per mm2 of Krt14+ medullary areas in indicated thymus graft sections. (I) Immunofluorescence analysis of tdTomato (red) and Krt14 (white) in indicated thymus graft sections. Scale bar, 50 μm. (J) Numbers of tdTomato+ cells per mm2 of Krt14+ medullary areas in indicated thymus graft sections. All images are representative data from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant.

Purity of isolated TECs for reaggregation with RelB-KO thymus stroma.
Shown are flow cytometric profiles of indicated cells from Ccl21atdTomato/+ E17 mice. Numbers indicate frequency of cells within indicated areas.
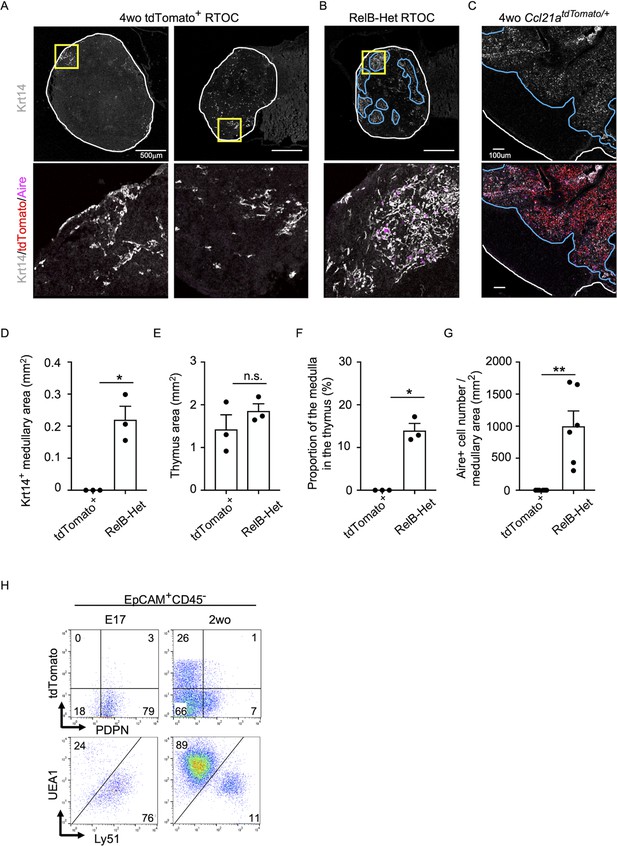
Developmental potential of postnatal CCL21-expressing medullary thymic epithelial cells (mTECs).
(A, B) Immunofluorescent staining of indicated thymus graft sections. Shown in upper panels are Krt14 expression (white). White lines indicate capsular outline of the thymus. Blue lines show Krt14+ medullary region. Scale bar, 500 μm. Bottom panels show Krt14 (white), tdTomato (red), and Aire (magenta) fluorescence signals within yellow boxes in upper panels. Grafts represent RelB-KO thymic stroma reaggregated with tdTomato+ TECs isolated from 4-week-old Ccl21atdTomato/+ mice (A) and RelB-heterozygous thymic stroma reaggregated without other TECs (B). (C) Immunofluorescent analysis of Krt14 (white), tdTomato (red), and Aire (magenta) in thymic sections from postnatal Ccl21atdTomato/+ mice. Scale bar, 100 μm. (D) Size of Krt14+ medullary areas (means and standard error of the means [SEMs], n = 3) in indicated thymus graft sections. (E) Size of grafted thymus areas (means and SEMs, n = 3) in indicated thymus graft sections. (F) Proportion (means and SEMs, n = 3) of Krt14+ medullary areas in the thymus areas in indicated thymus graft sections. (G) Numbers of Aire+ mTECs per mm2 of Krt14+ medullary areas in indicated thymus graft sections. All images are representative data from three independent experiments. *p < 0.05, **p < 0.01, n.s., not significant. (H) Flow cytometry profiles of tdTomato and podoplanin (PDPN) (top) and UEA1 and Ly51 (bottom) in EpCAM+CD45− TECs from Ccl21atdTomato/+ mice at E17 and 2 weeks old. Numbers indicate frequency of cells within indicated areas.
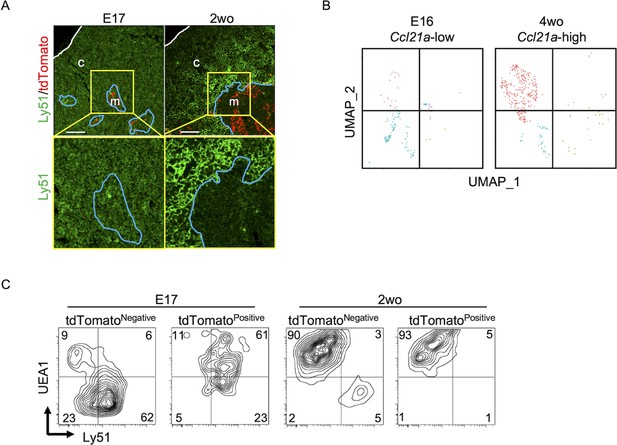
CCL21-expressing medullary thymic epithelial cells (mTECs) at E17 embryonic and postnatal period.
(A) Immunofluorescence analysis of thymus sections from E17 or 2-week-old Ccl21atdTomato/+ mice. tdTomato fluorescence (red) and Ly51 (green) expression are shown. White lines indicate thymic capsules. Blue lines indicate cortico-medullary junctions. Images in yellow boxes (top panels) are magnified in bottom panels. Representative data from at least three independent experiments are shown. c, cortex. m, medulla. Scale bar, 100 μm. (B) Uniform Manifold Approximation and Projection (UMAP) plots showing the clusters of Ccl21a+ mTECs in single-cell RNA-sequencing analysis of thymic epithelial cells isolated from E16 embryonic and 4-week-old postnatal mice. The majority of E16 Ccl21a+ mTECs were Ccl21alow, whereas the majority of postnatal Ccl21a+ mTECs were Ccl21ahigh (Figure 5B, C). (C) Flow cytometric profiles of UEA1 reactivity and Ly51 expression in tdTomatonegative EpCAM+CD45− TECs and tdTomatopositive EpCAM+CD45− TECs isolated from E17 embryonic and 2-week-old postnatal Ccl21atdTomato/+ mice. Numbers in the contour plots indicate frequency of cells within indicated area. Representative data from three independent experiments are shown.





