The evolution of transposable elements in Brachypodium distachyon is governed by purifying selection, while neutral and adaptive processes play a minor role
Figures
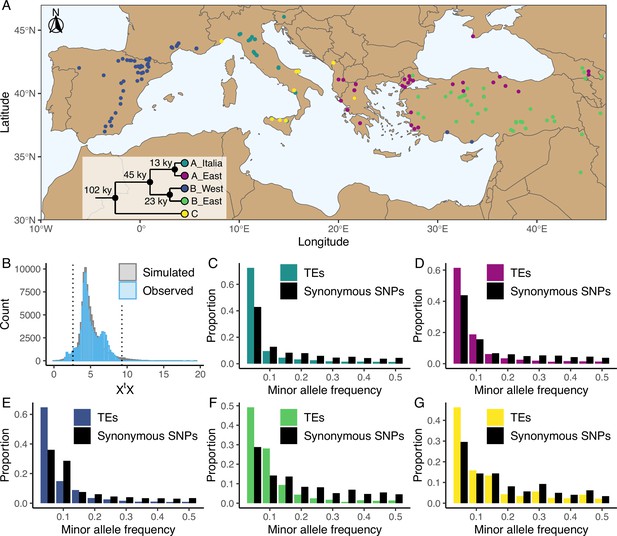
Distribution of the studied accessions and TE polymorphism frequencies.
(A) Map showing the geographical distribution of the accessions (n = 326) used in the current study. The phylogenetic tree illustrates the phylogeny between the five genetic clades. This panel was made based on the data and results published by Stritt et al., 2022 and Minadakis et al., 2023. (B) Observed (blue, n = 97,660) and simulated (gray, n = 100,000) XtX values of TE polymorphisms in B. distachyon. Dotted lines show the 2.5% and 97.5% quantiles of the simulated XtX values. (C-G) Folded site frequency spectrum of TE polymorphisms and synonymous SNPs in all clades. (C) A_East (nTE = 37,563; nSNP = 92,130); (D) A_Italia (nTE = 32,753; nSNP = 82,101); E: B_West (nTE = 48,315; nSNP = 99,953); F: B_East (nTE = 25,757; nSNP = 60,539); G: C (nTE = 24,161 ; nSNP = 78,681). Principal Component Analyses using TE, SNP, retrotransposon and DNA-transposon are shown in Figure 1—figure supplements 1 and 2. Observed correlation between age in generations and frequency of synonymous SNPs in the four derived genetic clades are shown in Figure 1—figure supplement 3. Distribution of the observed TE age scaled by the effective population size (Ne) in the four derived genetic clades are shown in Figure 1—figure supplement 4. Folded site frequency spectrum of DNA-transposons and retrotransposons are shown in Figure 1—figure supplements 5 and 6.
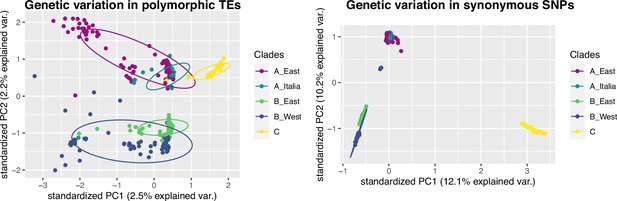
Principal Component Analyses using TE (left panel, n = 97,660) and SNP (right panel, n = 182,801) polymorphisms.
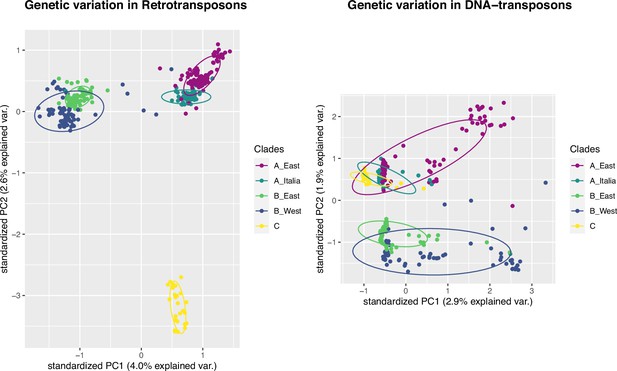
Principal Component Analyses using retrotransposon (left panel, n = 9,172) and DNA-transposon (right panel, n = 52,249) polymorphisms.

Observed correlation between age in generations and frequency of synonymous SNPs in the four derived genetic clades.
The red points show the expected age of a neutrally evolving mutation at a specific frequency based on the predictions of Kimura and Ohta, 1973. (A) A_East (n = 48,604); (B) A_Italia (n = 36,881); (C) B_West (n = 64,794) and (D) B_East (n = 36,892).
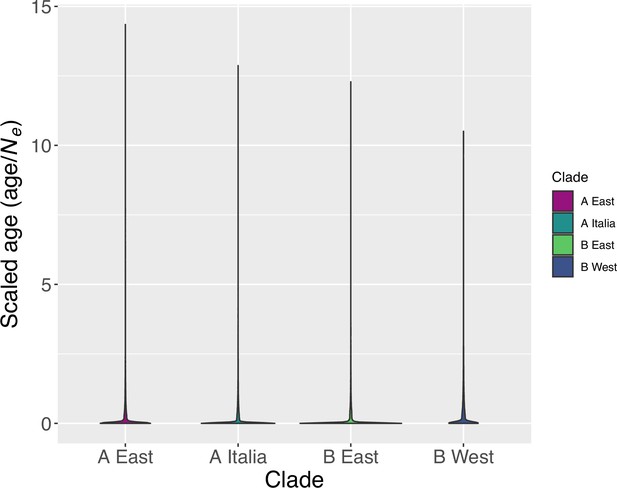
Distribution of the observed TE age scaled by the effective population size (Ne) in the four derived genetic clades of B. distachyon.
The age estimates were scaled by the effective population size to improve readability (n = 28,650; 13,867; 15,683; 26,672; respectively).
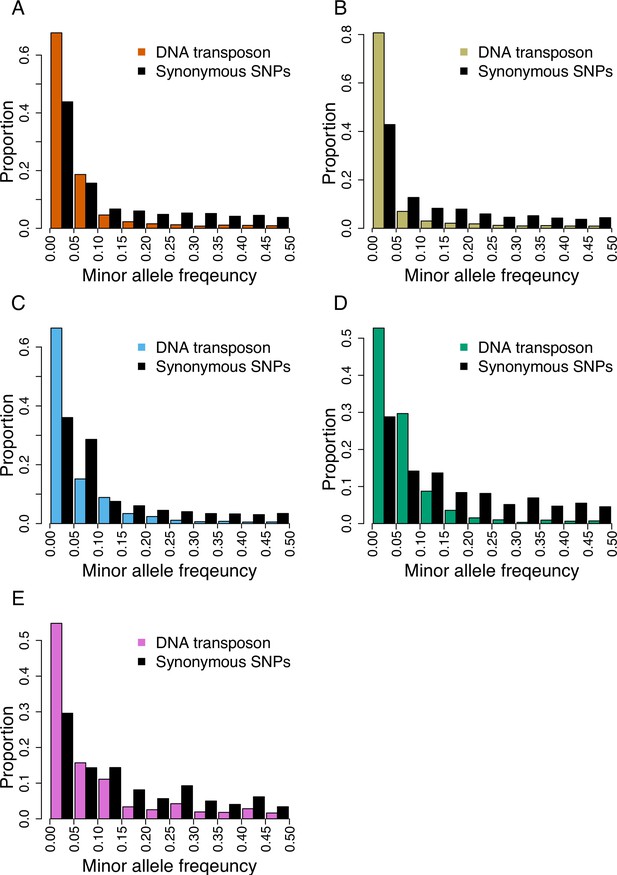
Folded site frequency spectrum of DNA-transposons and synonymous SNPs in all genetic clades.
Panel (A) A_East (nTE = 20,206 ; nSNP = 92,130); (B) A_Italia (nTE = 16,801 ; nSNP = 82,101); (C) B_West (nTE = 27,603 ; nSNP = 99,953); (D) B_East (nTE = 15,693 ; nSNP = 60,539); (E) C (nTE = 10,948 ; nSNP = 78,681).
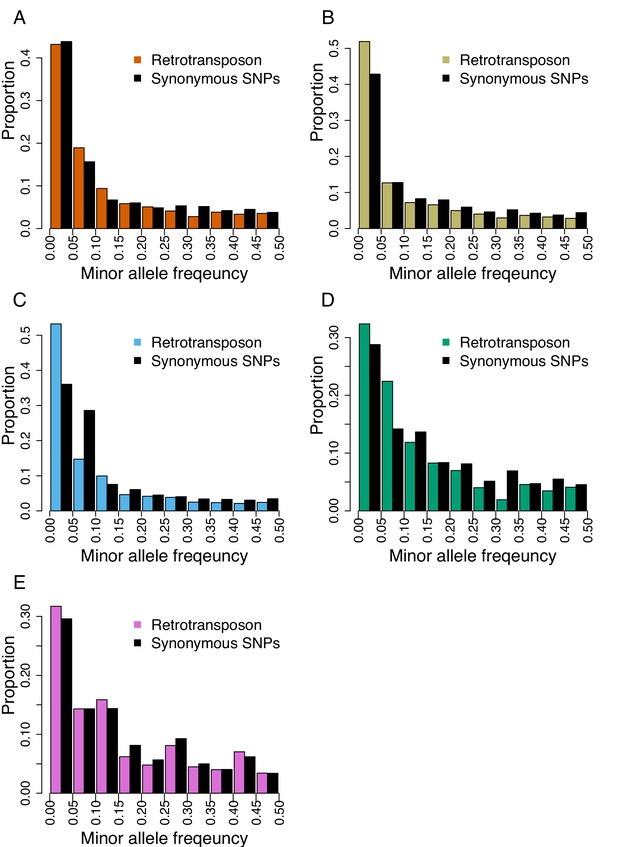
Folded site frequency spectrum of retrotransposons and synonymous SNPs in all genetic clades.
(A) A_East (nTE = 3,677 ; nSNP = 92,130); (B) A_Italia (nTE = 3,589 ; nSNP = 82,101); (C) B_West (nTE = 4,590 ; nSNP = 99,953); (D) B_East (nTE = 2,537 ; nSNP = 60,539); (E) C (nTE = 2,897 ; nSNP = 78,681).
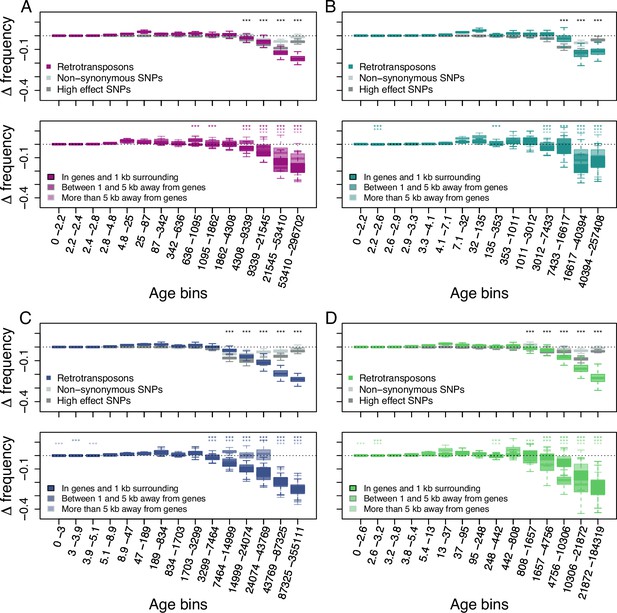
Age-adjusted SFS of retrotransposons.
The top row shows the age-adjusted SFS of all retrotransposons (colored), non-synonymous SNPs (light gray) and high effect SNPs (dark gray) in the four derived clades. The bottom row shows the age-adjusted SFS of retrotransposons based on their distance to the next gene in the four derived clades. The X axes show the age range of the mutations in each bin, and the age range of each bin was chosen so that each bin represents the same number of retrotransposon observations in the top row. The different columns show the four derived clades: (A) A_East (nretrotransposon = 2,106, nnon-synonymous SNP = 10,000, nhigh effect SNP = 9,050, nretrotransposon in genes and 1 kb surrounding = 733, nretrotransposon between 1 and 5 kb away from genes = 664, nretrotransposon more than 5 kb away from genes = 709); (B) A_Italia (nretrotransposon = 1,232, nnon-synonymous SNP = 10,000, nhigh effect SNP = 7,273, nretrotransposon in genes and 1 kb surrounding = 390, nretrotransposon between 1 and 5 kb away from genes = 388, nretrotransposon more than 5 kb away from genes = 454); (C) B_West (nretrotransposon = 2,081, nnon-synonymous SNP = 10,000, nhigh effect SNP = 10,000, nretrotransposon in genes and 1 kb surrounding = 812, nretrotransposon between 1 and 5 kb away from genes = 647, nretrotransposon more than 5 kb away from genes = 622); (D) B_East (nretrotransposon = 1,035 , nnon-synonymous SNP = 10,000, nhigh effect SNP = 6,306, nretrotransposon in genes and 1 kb surrounding = 387, nretrotransposon between 1 and 5 kb away from genes = 311, nretrotransposon more than 5 kb away from genes = 337). Boxplots are based on 100 estimations of D frequency. Significant deviations of D frequency estimates from 0 in the age-adjusted SFS of retrotransposons are shown with asterisks (one-side Wilcoxon tests, Bonferroni corrected p-value <0.01: ***). Age-adjusted SFS of DNA-transposons are shown in Figure 2—figure supplement 1. Age-adjusted SFS of simulated mutations under negative selection in the four derived clades transposons are shown in Figure 2—figure supplement 2. Age-adjusted SFS of retrotransposons in accessions with at least 20 x coverage are shown in Figure 2—figure supplement 3. Age-adjusted SFS of retrotransposons more than 5 kb away from genes are shown in Figure 2—figure supplement 4. Age-adjusted SFS of Copia, Ty3, Helitron and MITE TEs are shown in Figure 2—figure supplements 5–8.
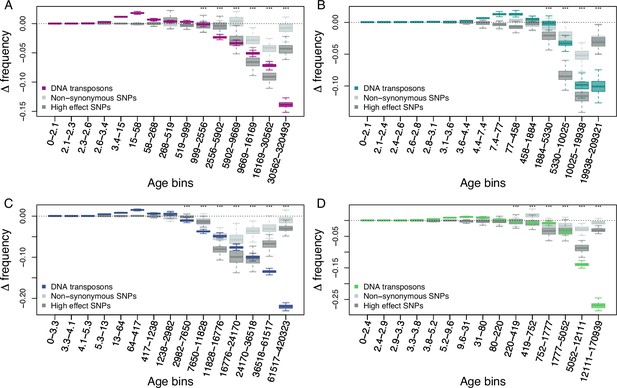
Age-adjusted SFS of DNA-transposons (colored), non-synonymous SNPs (light gray) and high effect SNPs (dark gray) in the four derived clades.
The X axes show the age range of the mutations in each bin, and the age range of each bin was chosen so that each bin represents the same number of DNA-transposons observations. (A) A_East (nDNA-transposon = 17,053, nnon-synonymous SNP = 10,000, nhigh effect SNP = 9,050); (B) A_Italia (nDNA-transposon = 7,538, nnon-synonymous SNP = 10,000, nhigh effect SNP = 9,050); (C) B_West (nDNA-transposon = 16,335, nnon-synonymous SNP = 10,000, nhigh effect SNP = 9,050); (D) B_East (nDNA-transposon = 10,101, nnon-synonymous SNP = 10,000, nhigh effect SNP = 9,050). Boxplots are based on 100 estimations of Δ frequency. Significant deviations of Δ frequency estimates from 0 in the age-adjusted SFS of DNA-transposons are shown with asterisks (one-side Wilcoxon tests, Bonferroni corrected p-value <0.01: ***).

Age-adjusted SFS of simulated mutations under negative selection in the four derived clades.
The four columns show the results for the A_East, A_Italia, B_West and B_East genetic clades, respectively. Each line shows the results for the different scaled selection coefficients (S). The five colored curves in each plot show the shape of the age-adjusted SFS with varying ratios of neutrally evolving mutations, and the gray curves show variation within one standard deviation based on the 20 runs for each simulation. The X axes show the age bin from the youngest to the oldest, with each age bin including the same number of observations for each simulation.
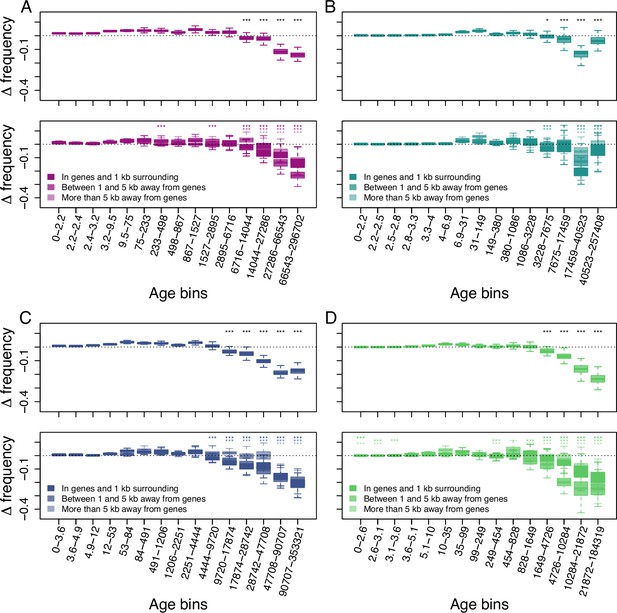
Age-adjusted SFS of retrotransposons in accessions with at least 20 x coverage.
The top row shows the age-adjusted SFS of retrotransposons in the four derived clades. The bottom row shows the age-adjusted SFS of retrotransposons based on their distance to the next gene in the four derived clades. The X axes show the age range of the mutations in each bin, and the age range of each bin was chosen so that each bin represents the same number of retrotransposon observations in the top row. The different columns show the four derived clades: (A): A_East (nretrotransposon = 1,688, nretrotransposon in genes and 1 kb surrounding = 564, nretrotransposon between 1 and 5 kb away from genes = 536, nretrotransposon more than 5 kb away from genes = 590); (B): A_Italia (nretrotransposon = 1,216, nretrotransposon in genes and 1 kb surrounding = 384, nretrotransposon between 1 and 5 kb away from genes = 381, nretrotransposon more than 5 kb away from genes = 451); (C): B_West (nretrotransposon = 1,911, nretrotransposon in genes and 1 kb surrounding = 746, nretrotransposon between 1 and 5 kb away from genes = 593, nretrotransposon more than 5 kb away from genes = 572); (D): B_East (nretrotransposon = 1,035, nretrotransposon in genes and 1 kb surrounding = 387, nretrotransposon between 1 and 5 kb away from genes = 311, nretrotransposon more than 5 kb away from genes = 337). Boxplots are based on 100 estimations of Δ frequency. Significant deviations of Δ frequency estimates from 0 in the age-adjusted SFS of retrotransposons are shown with asterisks (one-side Wilcoxon tests, Bonferroni corrected p-value <0.05: *;<0.01: ***).
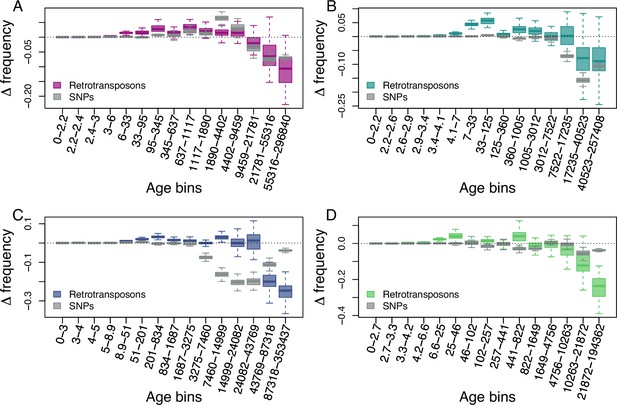
Age-adjusted SFS of retrotransposons (colored) and SNPs (gray) more than 5 kb away from genes in the four derived clades.
The X axes show the age range of the mutations in each bin. (A): A_East (nretrotransposon = 709); (B): A_Italia (nretrotransposon = 454); (C): B_West (nretrotransposon = 622); (D): B_East (nretrotransposon = 337). Boxplots are based on 100 estimations of Δ frequency.

Age-adjusted SFS of Copia TEs in the four derived clades.
The X axes show the age range of the mutations in each bin. (A): A_East (n = 1,027); (B): A_Italia (n = 621); (C): B_West (n = 1,066); (D): B_East (n = 531). Boxplots are based on 100 estimations of Δ frequency.

Age-adjusted SFS of Ty3 TEs in the four derived clades.
The X axes show the age range of the mutations in each bin. (A): A_East (n = 786)s; (B): A_Italia (n = 457); (C): B_West (n = 727); (D): B_East (n = 373). Boxplots are based on 100 estimations of Δ frequency.
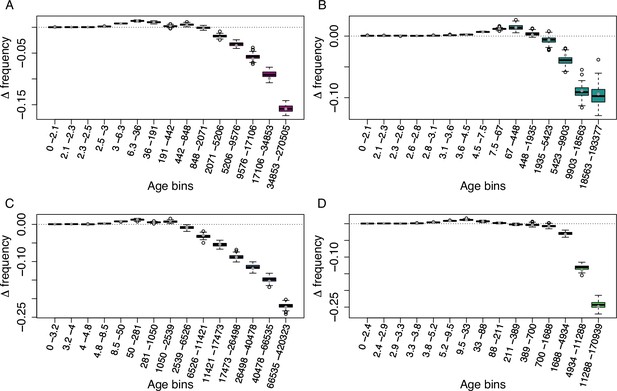
Age-adjusted SFS of Helitron TEs in the four derived clades.
The X axes show the age range of the mutations in each bin. (A): A_East (n = 8,895); (B): A_Italia (n = 4,291); (C): B_West (n = 8,324); (D): B_East (n = 5,736). Boxplots are based on 100 estimations of Δ frequency.
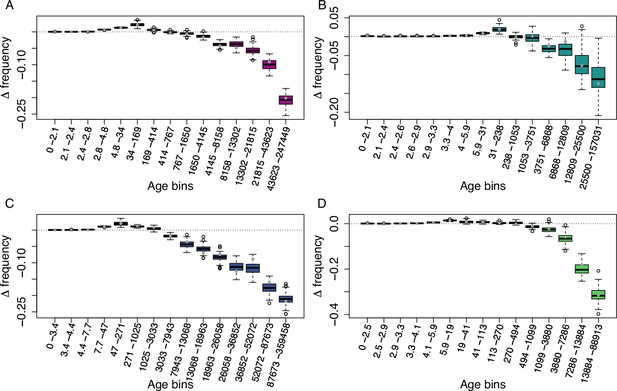
Age-adjusted SFS of MITE TEs in the four derived clades.
The X axes show the age range of the mutations in each bin. (A): A_East (n = 2,802); (B): A_Italia (n = 956); (C): B_West (n = 2,521); (D): B_East (n = 1,100). Boxplots are based on 100 estimations of Δ frequency.
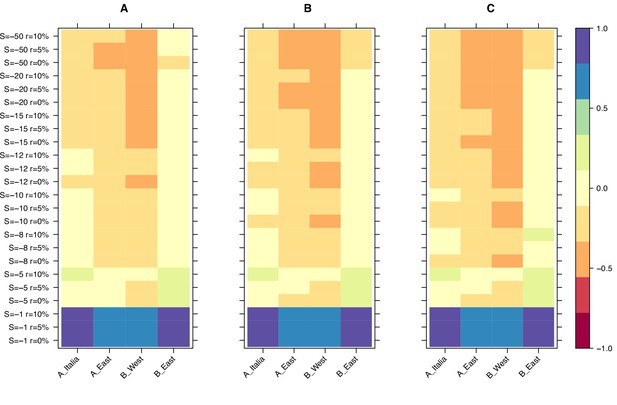
Relative age difference ((mutation age in simulations - observed mutation age)/maximum absolute age difference) between simulated and observed data in the last bin of the age-adjusted SFS.
(A): 25% quantile; (B): 50% quantile; (C): 75% quantile. Relative age difference between simulated data assuming fully outcrossing individuals and observed data in the last bin of the age-adjusted SFS are shown in Figure 3—figure supplement 1.
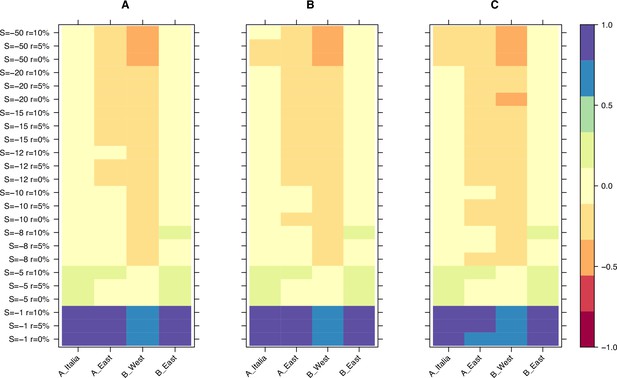
Relative age difference ((mutation age in simulations - observed mutation age)/maximum absolute age difference) between simulated data assuming fully outcrossing individuals and observed data in the last bin of the age-adjusted SFS.
(A): 25% quantile; (B): 50% quantile; (C): 75% quantile.
Tables
ANCOVA predicting the number of fixed TE polymorphisms per clade in candidate regions under positive selection.
| Variable | Sum of squares | degrees of freedom | F value | p value |
|---|---|---|---|---|
| Total number of TEs in the region | 28969.6 | 1 | 35405.64 | <0.001 |
| TE superfamily | 887.5 | 14 | 77.48 | <0.001 |
| Clade | 587 | 3 | 239.13 | <0.001 |
| Genomic region | 136.7 | 80 | 2.09 | <0.001 |
| TE age | 45.5 | 2 | 27.81 | <0.001 |
| High iHS | 0 | 1 | 0.03 | 0.869 |
ANCOVA predicting the allele frequency of TE polymorphisms per clade in candidate regions under positive selection.
| Variable | Sum of squares | degrees of freedom | F value | p value |
|---|---|---|---|---|
| TE superfamily | 453.2 | 14 | 247.3 | <0.001 |
| Clade | 17.7 | 3 | 45.18 | <0.001 |
| Genomic region | 147 | 80 | 14 | <0.001 |
| TE age | 2 | 2 | 7.7 | <0.001 |
| High iHS | 0.1 | 1 | 0.79 | 0.374 |
Additional files
-
Supplementary file 1
Supplementary tables.
Table 1a ANCOVA predicting the number of fixed TE polymorphisms per clade in candidate regions under positive selection in accessions with at least 20 x coverage. Table 1b ANCOVA predicting the allele frequency of TE polymorphisms per clade in candidate regions under positive selection in accessions with at least 20 x coverage. Table 1c List of TEs significantly associated with at least one environmental factor in the GWAS. The “Gene in the proximity” columns include information on the genes in the proximity of the TE insertion (less than 2 kb up and downstream). The last five columns indicate the frequency of the TE in each clade. Table 1d Difference in delta frequency between the oldest and second oldest age bin in the different simulations. Table 1e List of published samples used in this study. Because the reference accession was sequenced in multiple study and some samples were identified as outliers (indicating a wrong species classification or hybrid individuals) using PCA analyses on SNP and TE calls, only the samples listed below from each previous study were used. Table 1f List of thresholds used and percentage of the genome classified as high iHS regions in the four derived clades.
- https://cdn.elifesciences.org/articles/93284/elife-93284-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93284/elife-93284-mdarchecklist1-v1.pdf





