A single pair of pharyngeal neurons functions as a commander to reject high salt in Drosophila melanogaster
Figures
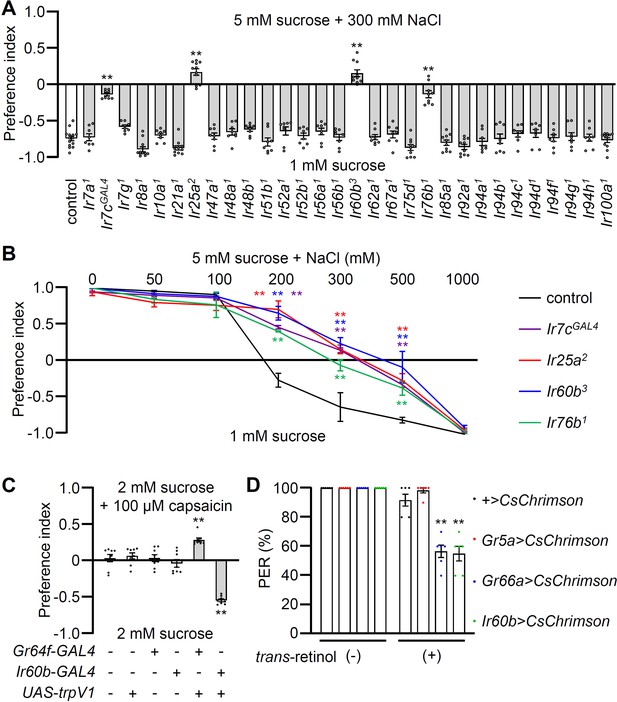
Testing requirements for Irs for avoiding high salt-containing food, and chemogenetic and optogenetic control of Ir60b gustatory receptor neurons (GRNs).
(A) Binary food choice assays (1 mM sucrose versus 5 mM sucrose and 300 mM NaCl) comparing 30 Ir mutants to the control strain (w1118) for high salt avoidance, n=8–12. (B) Preferences of the indicated flies for 1 mM sucrose versus 5 mM sucrose and 0–1000 mM NaCl. n=8–12. (C) Testing the effects of 100 μM capsaicin after expressing the rat TRPV1 channel (UAS-trpV1) either in Class A GRNs or Ir60b GRNs under the control of the Gr64f-GAL4 or the Ir60b-GAL4, respectively. The flies were given a choice between 2 mM sucrose and 2 mM sucrose plus 100 μM capsaicin. The presence or absence of the various transgenes is indicated by ‘+’ and ‘-’, respectively. n=8. (D) Testing the effects of light activation of various classes of GRNs. UAS-CsChrimson was expressed in Class A GRNs (driven by the Gr5a-GAL4), Class B GRNs (driven by the Gr66a-GAL4), or in Ir60b GRNs (driven by the Ir60b-GAL4). The flies were then simultaneously exposed to red lights (650 nm; WP-5700, 3M, USA) for 5 s while 2% sucrose was applied to labellum and the percent proboscis extension response (PER) was recorded. n=6. Data were compared using single-factor ANOVA coupled with Scheffe’s post-hoc test. Statistical significance was compared with the control. Means ± SEMs. **p<0.01.
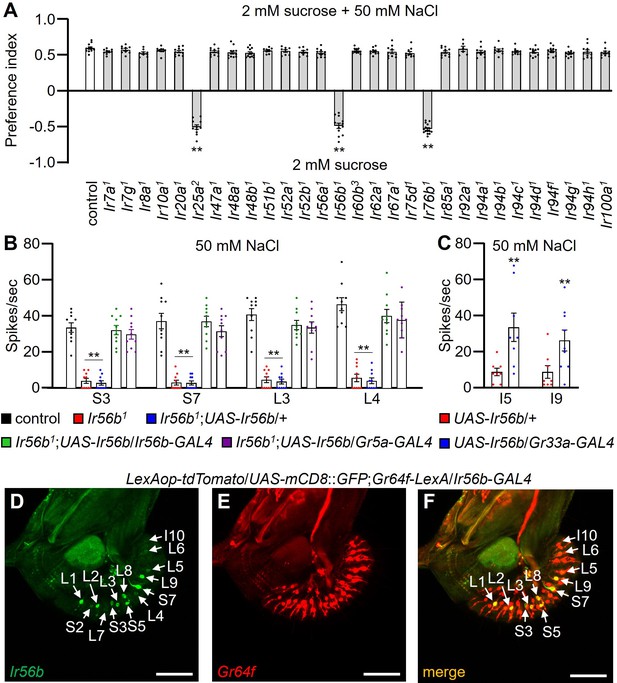
Requirements for Irs for preferring low salt-containing food.
(A) Binary food choice assays to assess low salt attraction by comparing Ir mutants with the control (w1118). n=8–12. (B) Tip recordings were performed on S3, S7, L3, and L4 sensilla. Shown are comparisons between control, Ir56b1, and flies expressing Ir56b under control of the Ir56b-GAL4 or the Gr5a-GAL4. n=8–12. (C) UAS-Ir56b expressed in Class B gustatory receptor neurons (GRNs) using the Gr33a-GAL4 driver. Tip recordings were conducted on I5 and I9 sensilla of the indicated flies with 50 mM NaCl. n=8. (D–F) Immunohistochemistry was performed using anti-GFP and anti-RFP on a labellum from a LexAop-tdTomato/UAS-mCD8::GFP;Gr64f-LexA/Ir56b-GAL4 fly. Multiple sets of data were compared using single-factor ANOVA coupled with Scheffe’s post-hoc test. Statistical significance was relative to the control. Means ± SEMs. **p<0.01. Scale bars in D–F indicate 20 μm.
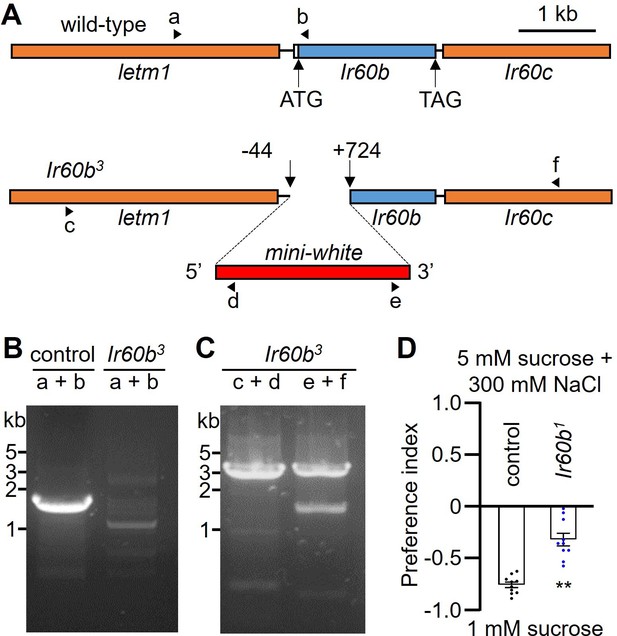
Gene structure of the Ir60b locus, generation of Ir60b3 and behavioral defect of Ir60b1 in high salt avoidance.
(A) Schematic of the Ir60b locus and Ir60b3 allele. The Ir60b coding exon is indicated by the blue rectangle. Ir60b3 was generated by ends-out homologous recombination by removing 768 base pairs as indicated. The red box indicates the insertion of the mini-white gene. The arrowheads (a–f) indicate the primers used for the PCR analyses in (B) and (C). (B) Confirmation of the deletion in Ir60b3 by PCR using primers a and b (primer a: 5'-TTGGTGTTTACTCGAAAACA-3', primer b: 5'-GCATTCAGAATGTATCTTAG-3'). (C) Confirmation of the deletion in Ir60b3 by PCR using primers c and d, and e and f (primer c: 5'-CGAACTGCATGCGCAACAGT-3', primer d: 5'-TTGCTGCCTCCGCGAATTAA-3', primer e: 5'-TGTACTACTCACATTGTTCA-3', primer f: 5'-GATTGTGAGCAGCAGCAGCA-3'). (D) Binary food choice assay testing control and Ir60b1 flies with 1 mM sucrose versus 5 mM sucrose and 300 mM NaCl. n=10. The pairwise comparison was conducted using a Student’s t-test. Means ± SEMs. **p<0.01.
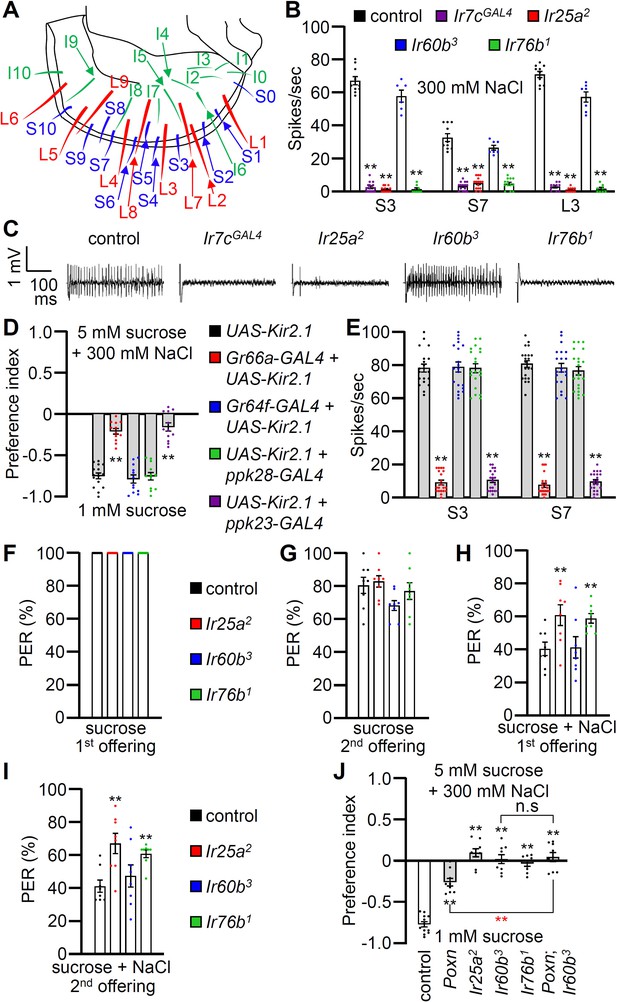
Contributions of different classes of gustatory receptor neurons (GRNs) to high salt avoidance.
(A) Schematic showing the names of sensilla bristles on the labellum Weiss et al., 2011. (B) Tip recordings conducted on S3, S7, and L3 sensilla using 300 mM NaCl and the indicated flies. n=10–16. (C) Representative traces obtained from S3 sensilla. (D) Binary food choice assays (1 mM sucrose versus 5 mM sucrose and 300 mM NaCl) after inactivating different classes of GRNs with UAS-Kir2.1, driven by the indicated GAL4 drivers: Class A (Gr64f; blue), Class B (Gr66a; red), Class C (ppk28; green), and Class D (ppk23; purple). Significances were determined by comparing to the UAS-Kir2.1 only control (black). n=12. (E) Tip recordings conducted by stimulating S3 and S7 sensilla with 300 mM NaCl from flies with different classes of GRNs inactivated with UAS-Kir2.1. See panel (D) for legend. n=16–20. (F–I) Proboscis extension response (PER) assays performed using the control strain (w1118; black) and Ir25a2 (red), Ir60b3 (blue), Ir76b1 (green). n=8–10. (F) PER percentages induced by 2% sucrose (first offering). (G) PER percentages induced by 2% sucrose (second offering). (H) PER percentages induced by 2% sucrose with 300 mM NaCl (first offering). (I) PER percentages induced by 2% sucrose with 300 mM NaCl (second offering). (J) Binary food choice assays for 300 mM salt avoidance were conducted with the control strain, Poxn (Poxn70-28/Poxn∆M22-B5), Ir25a2, Ir60b3, Ir76b1, and Poxn;Ir60b3. n=9–12. Data were compared using single-factor ANOVA coupled with Scheffe’s post-hoc test. Statistical significances compared with the control flies or the Poxn mutant are denoted by black and red asterisks, respectively. Means ± SEMs. **p<0.01.
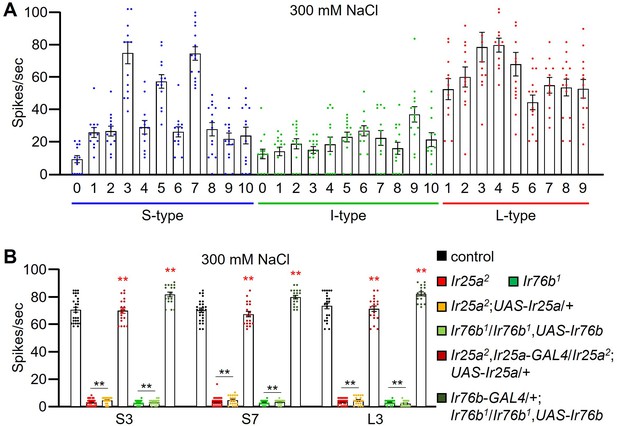
Assaying action potentials induced by different labellar bristles in response to 300 mM salt using tip recordings.
(A) Tip recordings were performed on S-type, I-type, and L-type sensilla from control flies. n=12–14. (B) Tip recordings conducted on S3, S7, and L3 sensilla from the indicated Ir25a2 and Ir76b1 mutants, as well as the mutants expressing wild-type UAS-Ir25a and UAS-Ir76b transgenes expressed under control of the Ir25a-GAL4 and the Ir76b-GAL4, respectively. n=16–28. Multiple sets of data were compared using single-factor ANOVA coupled with the Scheffe’s post-hoc test. Statistical significances compared to the control line are indicated by the black asterisks, while the red asterisks indicate significant rescue compared to the corresponding mutant. Means ± SEMs. **p<0.01.
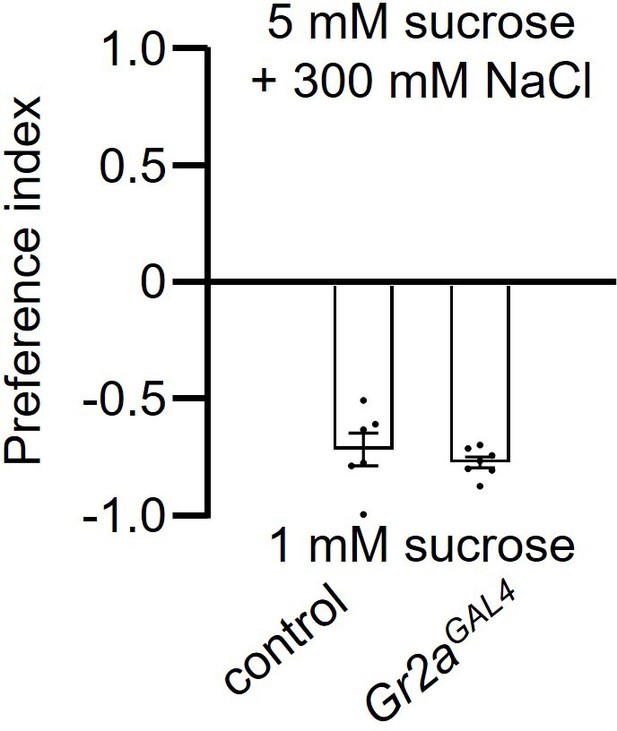
Two-way solid food choice assay to assess whether the Gr2aGAL4 mutant exhibits a deficit in avoidance of high salt.
The flies were given a choice between 1 mM sucrose versus 5 mM sucrose plus 300 mM NaCl in alternating wells of microtiter dishes. n=8. The pairwise comparison was conducted using an unpaired Student’s t-test. Means ± SEMs.
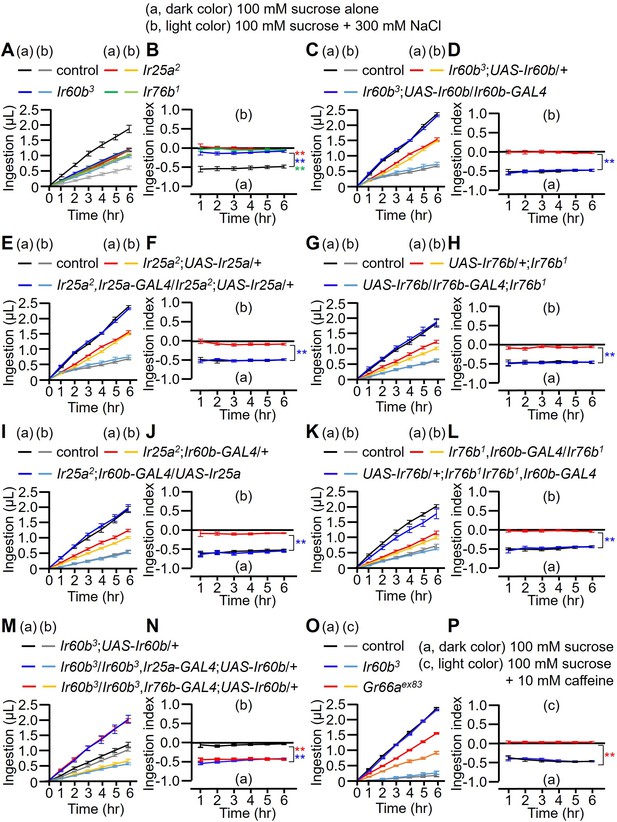
Measuring volume of food intake in Ir mutants using the DrosoX system.
(A–N) Each fly was exposed to two capillaries, one of which contained 100 mM sucrose (a), and the other contained 100 mM sucrose and 300 mM NaCl (b). (O and P) Each fly was exposed to two capillaries, one of which contained 100 mM sucrose (a), and the other contained 100 mM sucrose and 10 mM caffeine (c). (A, C, E, G, I, K, M, and O) Volumes of the two food options consumed by the indicated flies over the course of 6 hr. (B, D, F, H, I, J, L, N, and P) Ingestion indexes to indicate the relative consumption of the two foods. Ingestion indexes were calculated in each time point using the following equation: [(Ingestion volume of 100 mM sucrose and 300 mM NaCl or 10 mM caffeine) – (Ingestion volume of 100 mM sucrose)]/[(Ingestion volume of 100 mM sucrose and 300 mM NaCl or 10 mM caffeine) + (Ingestion volume of 100 mM sucrose)] n=12. Multiple sets of data were compared using single-factor ANOVA coupled with Scheffe’s post-hoc test. Statistical significances were relative to the control and determined for the ingestion indexes only. In all panels, the controls were w1118. The colors of the asterisks match the colors of the genotypes in the corresponding panels. Means ± SEMs. **p<0.01.
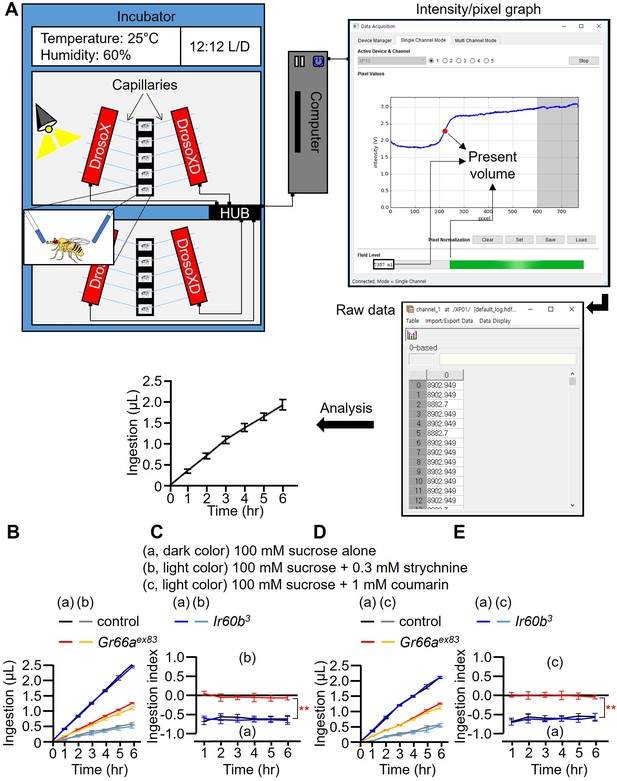
DrosoX system and measurement of food intake using strychnine and coumarin.
(A) The DrosoX system is composed of a cassette designed to accommodate five flies and a monitor that can hold five pairs of capillaries containing different solutions. Flies are exposed to both the DrosoX capillary and DrosoXD capillary options, enabling binary food (solution) choice experiments. A computer records the electrical signal generated by each photodiode, and the light intensity at each pixel in the array was sampled at 8 Hz by a microcontroller on a development board attached to the DrosoX sensor bank. Liquid level readings are acquired at sample rates of 2 Hz. The red dot on the blue line in the intensity/pixel graph on the right indicates the present volume. The raw data is displayed on the monitor with an accuracy of ±5.78 nL. The remaining volumes of solution are recorded based on the difference in optical density between air and the solution during the 6 hr experiment. The formula for calculating the ingestion volume is (Volumeinitial – Volumetime point). (B–E) Each fly (control, Ir60b3, and Gr66aex83) was exposed to two capillaries, one of which contained 100 mM sucrose (a), and the other contained 100 mM sucrose and either 0.3 mM strychnine or 1 mM coumarin as indicated (b). (B and D) Volumes of the two food options consumed by the indicated flies over the course of 6 hr. (C and E) Ingestion indexes (I.I.) to indicate the relative consumption of the two foods. I.I. formula: [Ingestion volume(b) or (c) – Ingestion volume(a)]/[Ingestion volume(b) or (c) + Ingestion volume(a)]. Multiple sets of data were compared using single-factor ANOVA coupled with the Scheffe’s post hoc test. n=12. Statistical significance compared with the controls is indicated by the asterisks. Means ± SEMs. **p<0.01.
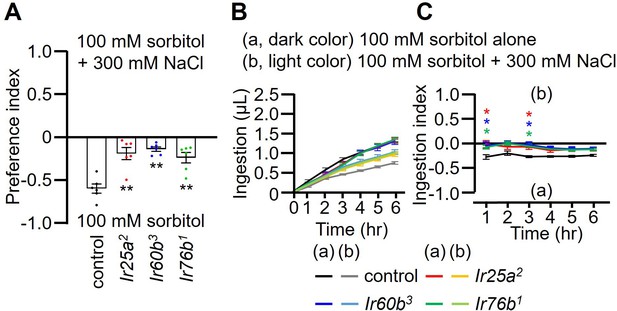
Two-way solid food choice assay and DrosoX binary capillary feeding assay using 100 mM sorbitol with or without 300 mM NaCl.
(A) Two-way solid food choice assay to assess whether the Ir25a2, Ir60b3, and Ir76b1 mutants exhibit a deficit in high salt avoidance. The flies were given a choice between 100 mM sorbitol and 100 mM sorbitol plus 300 mM NaCl in alternating wells of microtiter dishes. n=6. (B and C) DrosoX assays used to test the relative volumes consumed by control, Ir25a2, Ir60b3, and Ir76b1 flies when presented with capillaries containing either 100 mM sorbitol (a) or 100 mM sorbitol plus 300 mM NaCl (b). n=12. (B) Volumes of each of two food options. (C) Ingestion indexes (I.I.) to indicate the relative consumption of the two foods. I.I. formula: [Ingestion volume(b) – Ingestion volume(a)]/[Ingestion volume(b) + Ingestion volume(a)]. Multiple sets of data were compared using single-factor ANOVA coupled with Scheffe’s post-hoc test. Statistical significance compared with the controls. Means ± SEMs. *p<0.05. **p<0.01.
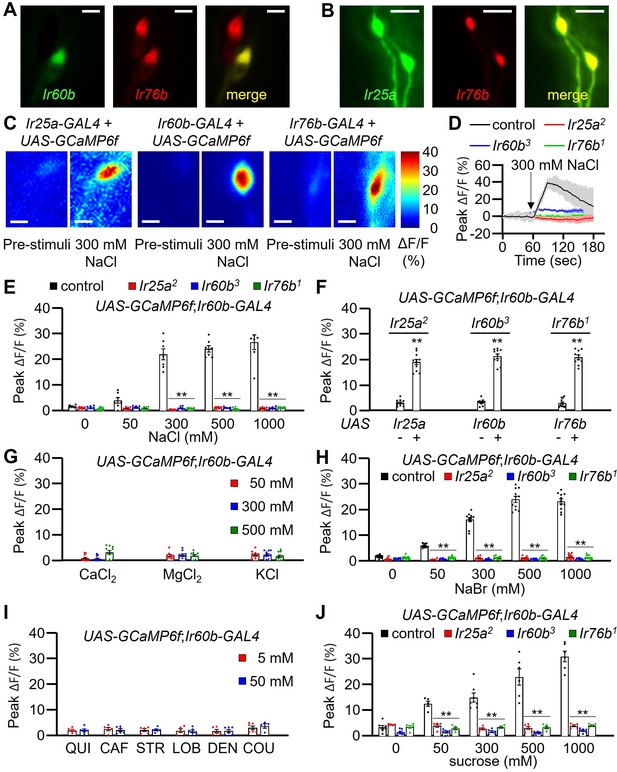
GCaMP6f responses of Ir60b gustatory receptor neurons (GRNs) to NaCl and other chemicals.
(A) Relative staining of the Ir60b reporter (green, anti-GFP) and the Ir76b reporter (red; anti-dsRed) in the labral sense organ (LSO) of UAS-mCD8::GFP/Ir76b-QF2;Ir60b-GAL4/QUAS-tdTomato flies. Merge is to the right. (B) Relative staining of the Ir25a reporter (green, anti-GFP) and the Ir76b reporter (red; anti-dsRed) in the LSO of Ir25a-GAL4/Ir76b-QF2;UAS-mCD8::GFP/QUAS-tdTomato. Merge is to the right. (C–J) Peak GCaMP6f responses (ΔF/F) of Ir60b GRNs in flies expressing UAS-GCaMP6f under control of the indicated GAL4 driver. (C) Heat map images illustrating changes in GCaMP6f fluorescence before and after stimulation with 300 mM NaCl using the indicated flies. (D) Sample traces depicting GCaMP6f responses to 300 mM NaCl. The traces are from the indicated flies expressing UAS-GCaMP6f driven by the Ir60b-GAL4. n=10–14. (E) GCaMP6f responses to various concentrations of NaCl in the indicated flies. UAS-GCaMP6f was driven by the Ir60b-GAL4. n=10–14. (F) GCaMP6f responses to 300 mM NaCl in the indicated mutants and in the absence or presence of the corresponding rescue transgene indicated by ‘-’ and ‘+’, respectively. n=8–10. (G) GCaMP6f responses to 50 mM, 300 mM, and 500 mM of CaCl2, MgCl2, and KCl in control flies. n=10–14. (H) GCaMP6f responses of Ir60b GRNs from the indicated flies to various concentrations of NaBr. n=10–14. (I) GCaMP6f responses to 5 mM and 50 mM concentrations of bitter compounds (quinine, caffeine, strychnine, lobeline, denatonium, and coumarin). n=8–10. (J) GCaMP6f responses to various concentrations of sucrose in Ir60b GRNs from the indicated flies. n=10–14. Multiple sets of data were compared using single-factor ANOVA coupled with Scheffe’s post-hoc test. Statistical significance compared with the controls. Means ± SEMs. **p<0.01. Scale bars in A–C indicate 5 μm.
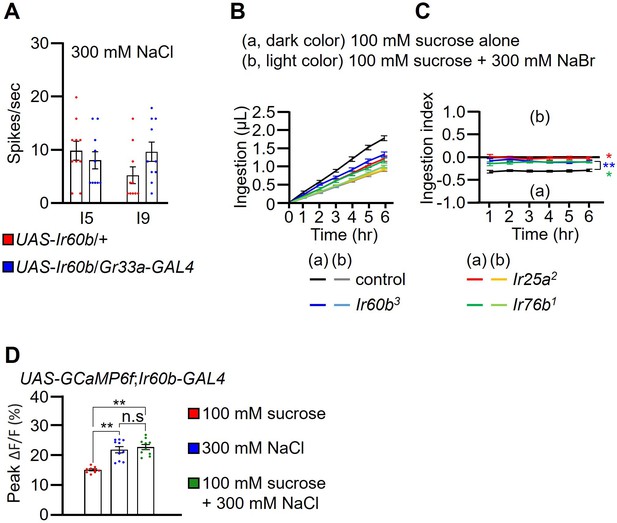
Testing whether ectopic expression of Ir60b confers responses to 300 mM NaCl, measurement of intake of sucrose plus 300 mM NaBr using the DrosoX assay, and Ca2+ response of Ir60b gustatory receptor neurons (GRNs).
(A) Tip recordings in response to 300 mM NaCl were performed on I5 and I9 sensilla of the indicated flies. UAS-Ir60b was expressed in Class B GRNs under control of the Gr33a-GAL4. n=10. (B and C) Droso-X assays used to test the relative volumes consumed by Ir25a2, Ir60b3, and Ir76b1 flies when presented with capillaries containing either 100 mM sucrose (a) or 100 mM sucrose plus 300 mM NaBr (b). n=12. (B) Volumes of each of two food options. (C) Ingestion indexes (I.I.) to indicate the relative consumption of the two foods. I.I. formula: [Ingestion volume(b) – Ingestion volume(a)]/[Ingestion volume(b) + Ingestion volume(a)]. (D) GCaMP6f responses of Ir60b GRNs to sucrose only, NaCl only, or a combination of sucrose and NaCl. n=10. Multiple sets of data were compared using single-factor ANOVA coupled with Scheffe’s post-hoc test. Statistical significance compared with the controls. Means ± SEMs. *p<0.05. **p<0.01.
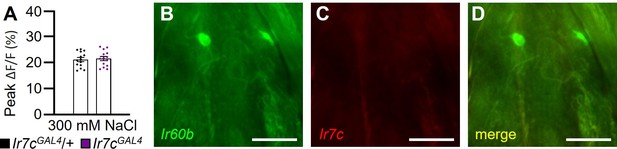
GCaMP6f responses evoked by 300 mM NaCl in Ir60b gustatory receptor neurons (GRNs) of control and Ir7cGAL4 flies, and relative expression of Ir60b and Ir7c reporters in the labral sense organ (LSO).
(A) GCaMP6f responses were stimulated by 300 mM NaCl in Ir60b GRNs from the Ir7cGAL4 mutant (Ir7cGAL4;UAS-GCaMP6f/+;Ir60b-GAL4/+) and from the Ir7cGAL4/+control (Ir7cGAL4/+;UAS-GCaMP6f/+;Ir60b-GAL4/+). n=8–10. The pairwise comparison was conducted using an unpaired Student’s t-test. Means ± SEMs. (B–D) Testing for expression of the Ir7c reporter in Ir60b GRNs. The Ir7c reporter consisted of a gene encoding GAL4 fused to RFP, and expressed under the direct control of the Ir7c promoter (Ir7cGAL4::VP166-RFP)1. The Ir60b reporter consisted of UAS-GFP driven by the Ir60b-GAL4. The genotype of the flies was Ir7cGAL4::VP166-RFP;Ir60b-GAL4;UAS-mCD8::GFP. The Ir60b reporter was detected with anti-GFP, and the Ir7c reporter was detected with anti-DsRed. Scale bars in B–D indicate 10 μm. Scale bars in B–D indicate 10 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-GFP | Molecular Probes | Cat # A11120; RPID: AB_221568 | IHC (1:1000) |
| Antibody | Rabbit polyclonal anti-DsRed | Clontech | Cat # 632496; RPID: AB_10013483 | IHC (1:1000) |
| Antibody | Goat polyclonal anti-mouse Alexa Fluro 488 | Invitrogen | Cat # A32723; RRID: AB_2633275 | IHC (1:200) |
| Antibody | Goat polyclonal anti-rabbit Alexa Fluor 568 | Invitrogen | Cat # A11011; RPID: AB_143157 | IHC (1:200) |
| Chemical compound | Sucrose | Sigma-Aldrich | Cat # 9378S | |
| Chemical compound | Tricholine citrate | Sigma-Aldrich | Cat # T0252 | |
| Chemical compound | Sulforhodamine B | Sigma-Aldrich | Cat # 230162 | |
| Chemical compound | Capsaicin | Sigma-Aldrich | Cat # M2028 | |
| Chemical compound | Caffeine | Sigma-Aldrich | Cat # C02750 | |
| Chemical compound | CaCl2 dihydrate | Sigma-Aldrich | Cat # C3881 | |
| Chemical compound | KCl | Sigma-Aldrich | Cat # P9541 | |
| Chemical compound | Quinine | Sigma-Aldrich | Cat # Q1125 | |
| Chemical compound | Strychnine | Sigma-Aldrich | Cat # S8753 | |
| Chemical compound | Lobeline | Sigma-Aldrich | Cat # 141879 | |
| Chemical compound | Denatonium | Sigma-Aldrich | Cat # D5765 | |
| Chemical compound | Coumarin | Sigma-Aldrich | Cat # C4261 | |
| Chemical compound | Brilliant blue FCF | Wako Pure Chemical Industry | Cat # 027-12842 | |
| Chemical compound | Paraformaldehyde | Electron Microscopy Sciences | Cat # 15710 | |
| Chemical compound | NaCl | LPS Solution | Cat # NACL01 | |
| Chemical compound | MgCl2 hexahydrate | SAMCHUN | Cat # M0038 | |
| Chemical compound | NaBr | DUKSAN | Cat # S2531 | |
| Chemical compound | Goat serum, New Zealand origin | Gibco | Cat # 16210064 | |
| Genetic reagent (Drosophila melanogaster) | w1118 | Bloomington Drosophila Stock Center (BDSC) | BDSC:5905 | |
| Genetic reagent (Drosophila melanogaster) | Ir7a1 | Dr. Y Lee Rimal et al., 2019 | ||
| Genetic reagent (Drosophila melanogaster) | Ir7g1: y1w*Mi{y+mDint2=MIC}Ir7gMI06687 | BDSC | BDSC:42420 | |
| Genetic reagent (Drosophila melanogaster) | Ir7cGAL4 | Dr. MD Gordon McDowell et al., 2022 | ||
| Genetic reagent (Drosophila melanogaster) | Ir8a1: w*TI{w[+m*]=TI}Ir8a1;Bl1L2/CyO | BDSC | BDSC:23842 | |
| Genetic reagent (Drosophila melanogaster) | Ir10a1: w1118Mi{GFPE.3xP3=ET1}Ir10aMB03273 | BDSC | BDSC:41744 | |
| Genetic reagent (Drosophila melanogaster) | Ir21a1: w1118;PBac{w+mC=PB}Ir21ac02720 | BDSC | BDSC:10975 | |
| Genetic reagent (Drosophila melanogaster) | Ir25a2 | Dr. L Vosshall Benton et al., 2009 | ||
| Genetic reagent (Drosophila melanogaster) | Ir47a1 | Dr. Y Lee Rimal et al., 2019 | ||
| Genetic reagent (Drosophila melanogaster) | Ir48a1: w1118;Mi{GFPE.3xP3= ET1}Ir48aMB09217 | BDSC | BDSC:26453 | |
| Genetic reagent (Drosophila melanogaster) | Ir48b1: w1118;Mi{GFPE.3xP3= ET1}Ir48bMB02315 | BDSC | BDSC:23473 | |
| Genetic reagent (Drosophila melanogaster) | Ir51b1: w1118;PBac{w+mC=PB}rowc00387 Ir51bc00387 | BDSC | BDSC:10046 | |
| Genetic reagent (Drosophila melanogaster) | Ir52a1 | Dr. Y Lee Rimal et al., 2019 | ||
| Genetic reagent (Drosophila melanogaster) | Ir52b1: w1118;Mi{GFPE.3xP3= ET1}Ir52bMB02231/SM6a | BDSC | BDSC:25212 | |
| Genetic reagent (Drosophila melanogaster) | Ir52c1: w1118;Mi{GFPE.3xP3= ET1}Ir52cMB04402 | BDSC | BDSC:24580 | |
| Genetic reagent (Drosophila melanogaster) | Ir56a1 | Dr. Y Lee Rimal et al., 2019 | ||
| Genetic reagent (Drosophila melanogaster) | Ir56b1: w1118;Mi{GFPE.3xP3=ET1}Ir56bMB09950 | BDSC | BDSC:27818 | |
| Genetic reagent (Drosophila melanogaster) | Ir56d1: w*;Ir56d1 | BDSC | BDSC:81249 | |
| Genetic reagent (Drosophila melanogaster) | Ir60b1 | Dr. J Carlson Joseph et al., 2017 | ||
| Genetic reagent (Drosophila melanogaster) | Ir60b3 | Dr. Y Lee | In this study | |
| Genetic reagent (Drosophila melanogaster) | Ir62a1: y1w*;Mi{y+mDint2=MIC} Ir62aMI00895Iml1MI00895/TM3, Sb1 Ser1 | BDSC | BDSC:32713 | |
| Genetic reagent (Drosophila melanogaster) | Ir67a1: y1w*;Mi{y+mDint2=MIC}Ir67aMI11288 | BDSC | BDSC:56583 | |
| Genetic reagent (Drosophila melanogaster) | Ir75d1: w1118;Mi{GFPE.3xP3=ET1}Ir75dMB04616 | BDSC | BDSC:24205 | |
| Genetic reagent (Drosophila melanogaster) | Ir76b1 | Dr. C Montell Zhang et al., 2013 | ||
| Genetic reagent (Drosophila melanogaster) | Ir85a1: w1118;Mi{GFPE.3xP3=ET1} Ir85aMB04613 Pif1AMB04613 | BDSC | BDSC:24590 | |
| Genetic reagent (Drosophila melanogaster) | Ir92a1: w1118;Mi{GFPE.3xP3=ET1}Ir92aMB03705 | BDSC | BDSC:23638 | |
| Genetic reagent (Drosophila melanogaster) | Ir94a1 | Dr. Y Lee Rimal et al., 2019 | ||
| Genetic reagent (Drosophila melanogaster) | Ir94b1:w1118;Mi{GFPE.3xP3= ET1}Ir94bMB02190 | BDSC | BDSC:23424 | |
| Genetic reagent (Drosophila melanogaster) | Ir94c1 | Dr. Y Lee Rimal et al., 2019 | ||
| Genetic reagent (Drosophila melanogaster) | Ir94d1:y1w*;Mi{y+mDint2=MIC} Ir94dMI01659CG17380MI01659 | BDSC | BDSC:33132 | |
| Genetic reagent (Drosophila melanogaster) | Ir94f1: y1w*;Mi{y+mDint2= MIC}Ir94fMI00928 | BDSC | BDSC:33095 | |
| Genetic reagent (Drosophila melanogaster) | Ir94g1: w1118;Mi{GFPE.3xP3= ET1}Ir94gMB07445 | BDSC | BDSC:25551 | |
| Genetic reagent (Drosophila melanogaster) | Ir94h1 | Dr. Y Lee Rimal et al., 2019 | ||
| Genetic reagent (Drosophila melanogaster) | Ir100a1: w1118;P{w+mC=EP} Ir100aG19846 CG42233G19846 | BDSC | BDSC:31853 | |
| Genetic reagent (Drosophila melanogaster) | UAS-mCD8::GFP | BDSC | BDSC:5137 | |
| Genetic reagent (Drosophila melanogaster) | UAS-mCD8::GFP | BDSC | BDSC:32184 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Kir2.1 | BDSC | BDSC:6595 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Ir25a | Dr. Y Lee Lee et al., 2018 | ||
| Genetic reagent (Drosophila melanogaster) | UAS-Ir60b | Dr. Y Lee | In this study | |
| Genetic reagent (Drosophila melanogaster) | UAS-Ir76b | Dr. C Montell Zhang et al., 2013 | ||
| Genetic reagent (Drosophila melanogaster) | Ir25a-GAL4 | Dr. L Vosshall Benton et al., 2009 | ||
| Genetic reagent (Drosophila melanogaster) | Ir60b-GAL4 | Dr. C Montell Joseph et al., 2017 | ||
| Genetic reagent (Drosophila melanogaster) | Ir76b-GAL4 | Dr. C Montell Zhang et al., 2013 | ||
| Genetic reagent (Drosophila melanogaster) | ppk23-GAL4 | Dr. K Scott Thistle et al., 2012 | ||
| Genetic reagent (Drosophila melanogaster) | ppk28-GAL4 | Dr. H Amrein Cameron et al., 2010 | ||
| Genetic reagent (Drosophila melanogaster) | Gr66a-GAL4 | Dr. H Amrein Thorne et al., 2004 | ||
| Genetic reagent (Drosophila melanogaster) | Gr64f-GAL4 | Dr. A Dahanukar Lee et al., 2018 | ||
| Genetic reagent (Drosophila melanogaster) | Ir76b-QF | BDSC | BDSC:51312 | |
| Genetic reagent (Drosophila melanogaster) | QUAS-tdTomato: y1w1118; P{QUAS-mtdTomato-3xHA}26 | BDSC | BDSC:30005 | |
| Genetic reagent (Drosophila melanogaster) | Poxn∆M22-B5: y1w67c23; Mi{ET1}PoxnMB00113 | BDSC | BDSC:22701 | |
| Genetic reagent (Drosophila melanogaster) | Poxn70-28: Poxn70/CyO; twi-Gal4, UAS-2XEGFP | BDSC | BDSC:60688 | |
| Software | Origin Pro Version | Dr. Y Lee | https://www.originlab.com | |
| Software | GraphPad Prism | Dr. Y Lee | https://www.graphpd.com | |
| Software | Autospike 3.1 software | Dr. Y Lee | https://www.syntech.co.za/ | |
| Software | Fiji/ImageJ software | Dr. Y Lee | https://fiji.sc | |
| Software | ZEN lite 2.5 blue | Dr. Y Lee | https://www.zeiss.com/ |





