A myristoyl switch at the plasma membrane triggers cleavage and oligomerization of Mason-Pfizer monkey virus matrix protein
Figures
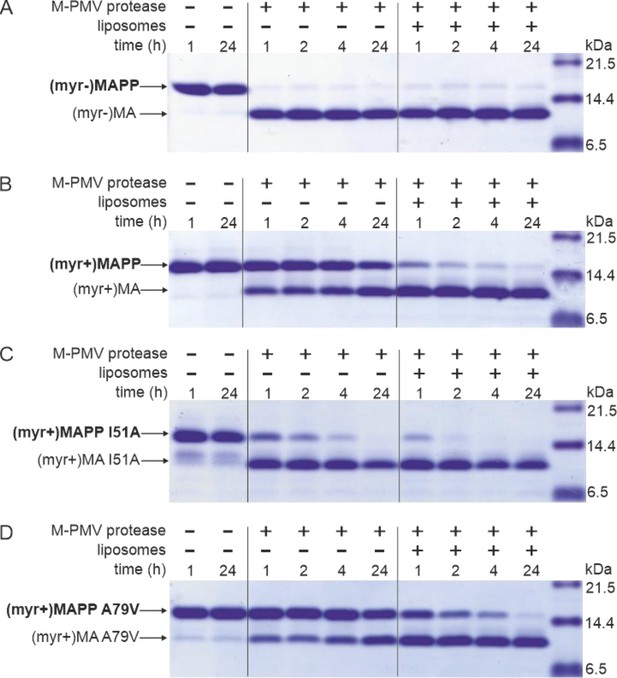
MAPP cleavage by Mason-Pfizer monkey virus (M-PMV) protease.
(A) (myr−)MAPP, (B) (myr+)MAPP, (C) ‘myrOUT’ mutant (myr+)MAPP I51A, and (D) ‘myrIN’ mutant (myr+)MAPP A79V were cleaved by M-PMV protease in the absence or presence of liposomes mimicking the plasma membrane (PM) inner leaflet. All the experiments were performed in three biological replicates with the same results. Source data – Figure 1—source data 1.
-
Figure 1—source data 1
Source data for Figure 1 containing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels.
- https://cdn.elifesciences.org/articles/93489/elife-93489-fig1-data1-v3.zip
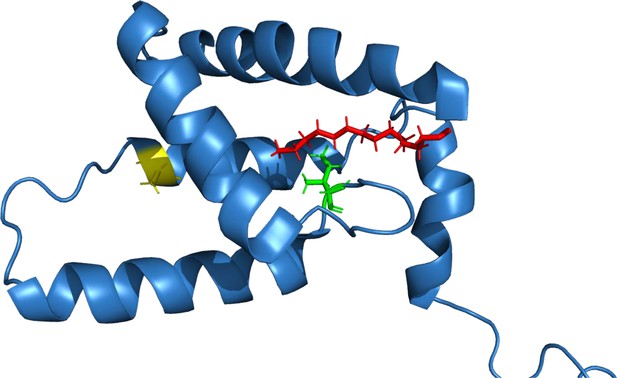
Structure of wild-type (WT) (myr+)MAPP indicating positions of the mutated residues.
The positions of mutated residues are indicated in previously published structure (Prchal et al., 2012): I51 (green) and A79 (yellow). Myristoyl is shown in red.
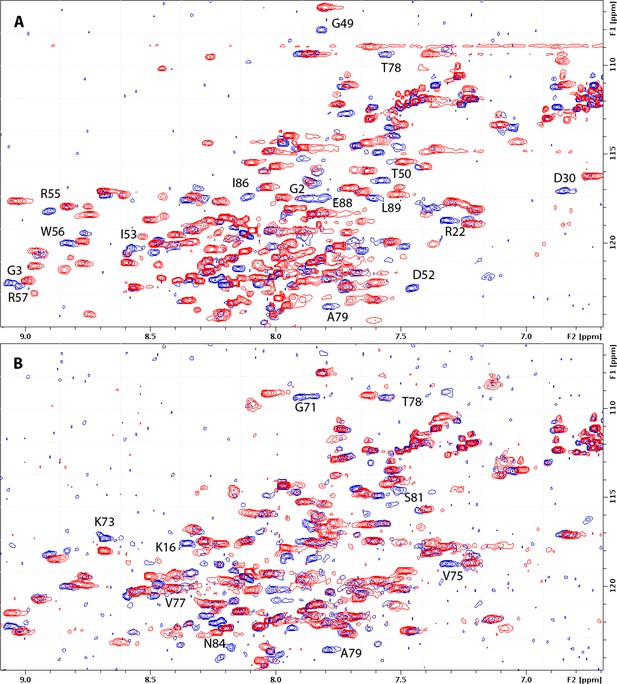
Comparison of (myr+)MAPP wild-type (WT) and mutants HN-HSQC (1H–15N-heteronuclear single quantum coherence) spectra.
HN-HSQC spectra of WT (myr+)MAPP (blue) overlaid with the same spectra of I51A (red, panel A) and A79V (red, panel B) mutants. Signals of WT HN groups that are not overlapped by signals of mutant are assigned.
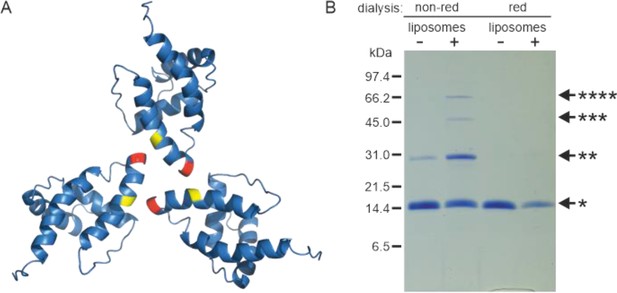
Oligomerization of (myr−)MAPP.
(A) Previously published structure of wild-type (WT) (myr−)MA trimer (Vlach et al., 2009) with residues 62 (yellow) and 69 (red) highlighted. (B) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel showing oligomers of T69C (myr+)MAPP formed upon the interaction of T69C (myr+)MAPP with liposomes, stabilized by disulfide bridges under non-reducing conditions. non-red – non-reducing conditions; red – reducing conditions; *monomer, **dimer, ***trimer, and ****tetramer. Source data – Figure 2—source data 1.
-
Figure 2—source data 1
Source data for Figure 2 containing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel.
- https://cdn.elifesciences.org/articles/93489/elife-93489-fig2-data1-v3.zip
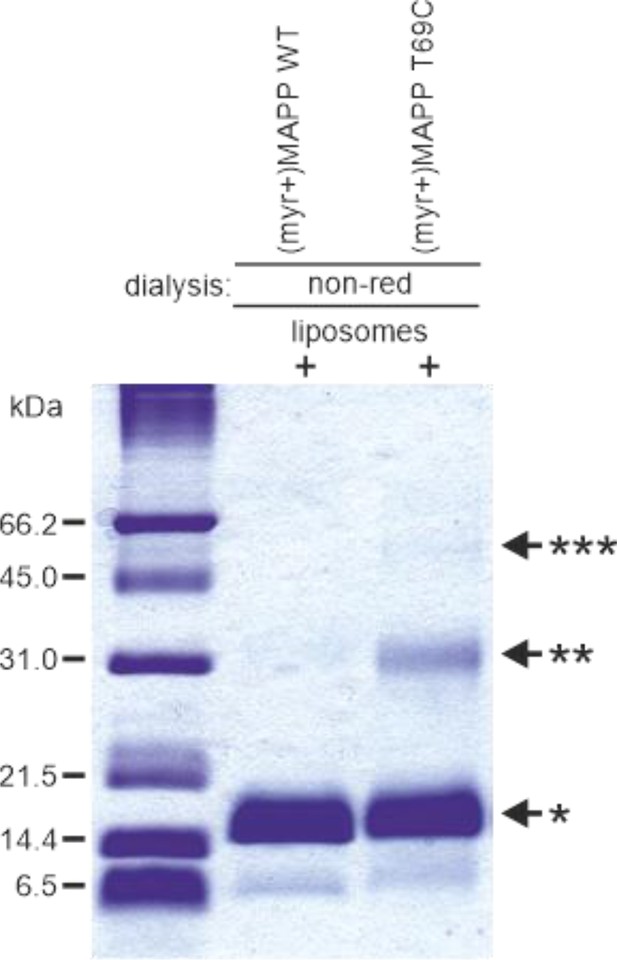
Oligomerization of (myr−)MAPP T69C.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel showing monomer of (myr+)MAPP wild-type (WT) and oligomers of (myr+)MAPP T69C formed upon the interaction with liposomes, stabilized by disulfide bridges under non-reducing conditions. non-red – non-reducing conditions; *monomer, **dimer, and ***trimer. Source data – Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1 containing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel.
- https://cdn.elifesciences.org/articles/93489/elife-93489-fig2-figsupp1-data1-v3.zip
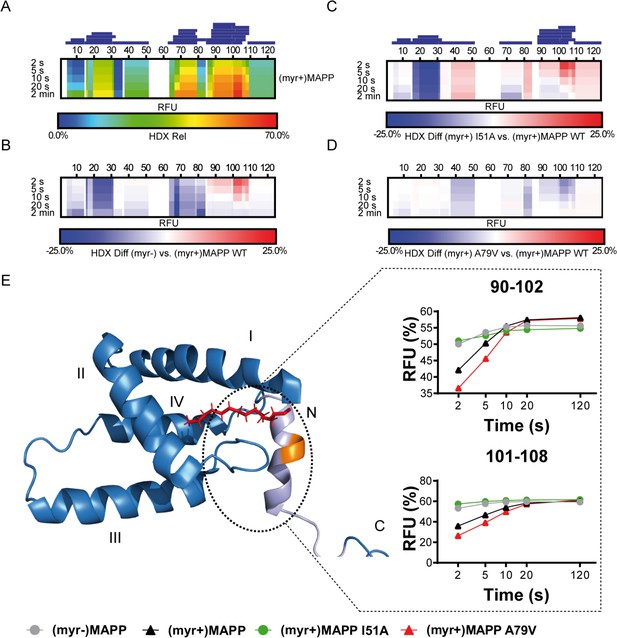
Deuterium exchange rates of (myr+)MAPP wild-type (WT) and its comparison to (myr−)MAPP WT and I51A and A79V mutants of (myr+)MAPP.
Comparison of hydrogen–deuterium exchange (HDX) rates of backbone amide hydrogens for (myr+)MAPP WT with HDX rates of other protein forms. Panels A–D: Heat maps show the HDX rate of identified peptides indicated as blue rectangles above the amino acid numbering. HDX is shown in rainbow colors (A) or in blue–white–red color scale for differential heat maps (B–D) as percent relative fractional uptake (RFU, %) at the corresponding time points. Regions without HDX data are shown as white spaces. (A) The HDX heat map of (myr+)MAPP WT. (B) Differential HDX heat map of (myr−)MAPP WT compared to (myr+)MAPP WT. (C) Differential HDX heat map of (myr+)MAPP I51A compared to (myr+)MAPP WT. (D) Differential HDX heat map of (myr+)MAPP A79V compared to (myr+)MAPP WT. In differential HDX maps the less accessible amino acids/regions are displayed in blue, while more accessible ones are in red. (E) The previously published NMR structure of (myr+)MAPP (Prchal et al., 2012, RCSB PDB: 5LMY) with protease cleavage site shown in orange, myristoyl in red, residues 96–110 in light violet, and first four helices of MA are numbered. Graphs show differences in HDX exchange rates in the 90–108 regions around the protease cleavage site represented by peptides 90–102 and 101–108 for different forms of MAPP protein. Related source data – Figure 3—source data 1, Figure 3—source data 2, and Figure 3—source data 3.
-
Figure 3—source data 1
Comparison of hydrogen–deuterium exchange (HDX) of (myr+)MAPP and (myr−)MAPP – data related to Figure 3B.
- https://cdn.elifesciences.org/articles/93489/elife-93489-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Comparison of hydrogen–deuterium exchange (HDX) of (myr+)MAPP and I51A (myr+)MAPP – data related to Figure 3C.
- https://cdn.elifesciences.org/articles/93489/elife-93489-fig3-data2-v3.xlsx
-
Figure 3—source data 3
Comparison of hydrogen–deuterium exchange (HDX) of (myr+)MAPP and A79V (myr+)MAPP – data related to Figure 3D.
- https://cdn.elifesciences.org/articles/93489/elife-93489-fig3-data3-v3.xlsx
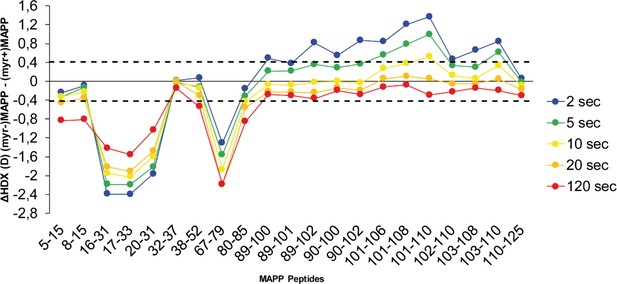
Comparison of hydrogen–deuterium exchange (HDX) of (myr+)MAPP and (myr−)MAPP wild types (WTs).
The butterfly plot shows the HDX differences between WT (myr+)MAPP and WT (myr−)MAPP over the time (see also Figure 3—source data 1). Peptides with significant differences in HDX are arranged according to their positions in MAPP. Confidence threshold (CI98%) is displayed as a dotted black line. The value of confidence threshold was calculated as ±0.410 Da.
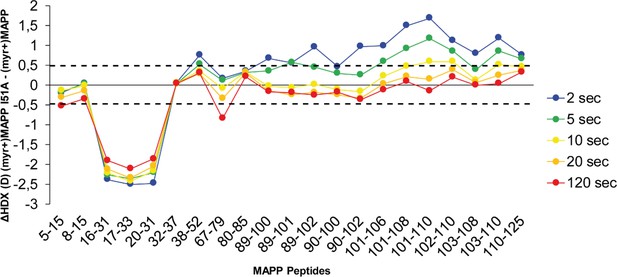
Comparison of hydrogen–deuterium exchange (HDX) of (myr+)MAPP and I51A (myr+)MAPP.
The butterfly plot shows the HDX differences between wild-type (WT) (myr+)MAPP and I51A (myr+)MAPP over the time (see also Figure 3—source data 2). Peptides with significant differences in HDX are arranged according to their positions in MAPP. Confidence threshold (CI98%) is displayed as a dotted black line. The value of confidence threshold was calculated as ±0.478 Da.
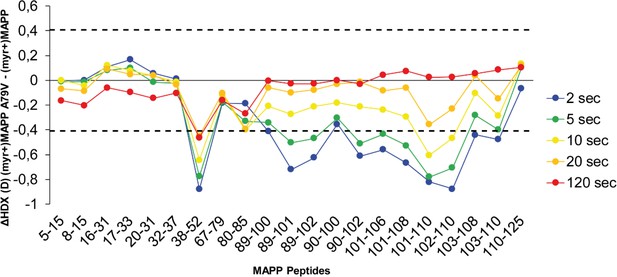
Comparison of hydrogen–deuterium exchange (HDX) of (myr+)MAPP and A79V (myr+)MAPP.
The butterfly plot shows the HDX differences between wild-type (WT) (myr+)MAPP and A79V (myr+)MAPP over the time (see also Figure 3—source data 3). Peptides with significant differences in HDX are arranged according to their positions in MAPP. Confidence threshold (CI98%) is displayed as a dotted black line. The value of confidence threshold was calculated as ±0.410 Da.
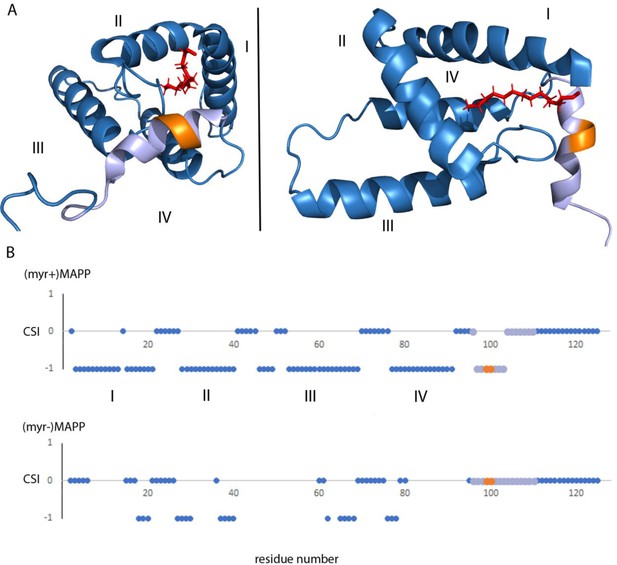
Structure of (myr+)MAPP (Prchal et al., 2012), position and secondary structure of the cleavage site between MA and PP.
(A) The protease cleavage site in (myr+)MAPP is shown in orange, myristoyl in red, residues 96–110 in light violet. The first four helices of MA (blue) are numbered, the myristoyl is shown in red. (B) Backbone chemical shifts analysis of (myr+)MAPP and (myr−)MAPP proteins using TALOS+ software. CSI (Chemical Shift Index) 0 indicates that residue is in the loop region, CSI −1 indicates that residue is in alpha-helical region. The protease cleavage site is shown in orange, residues 96–110 in light violet. The first four helices of MA (blue) are numbered.
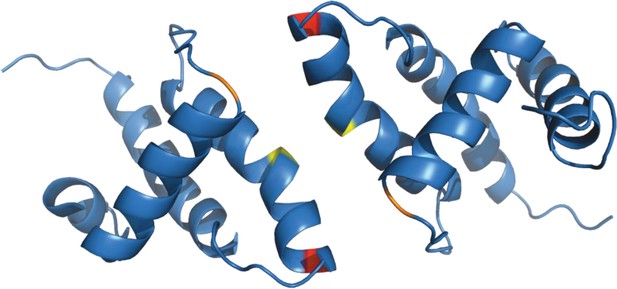
Structure of (myr−)MA wild-type dimer (Vlach et al., 2009).
Previously published ribbon structure of (myr−)MA wild-type dimer (Vlach et al., 2009) showing positions of residues T69 (in red), C62 (in yelow), and C42 (in orange).
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93489/elife-93489-mdarchecklist1-v3.pdf
-
Supplementary file 1
Comparison of observed and calculated backbone atoms chemical shifts for (myr+)MAPP and (myr-)MAPP.
- https://cdn.elifesciences.org/articles/93489/elife-93489-supp1-v3.xlsx
-
Supplementary file 2
Estimated backbone order parameters and secondary structure for (myr+)MAPP and (myr-)MAPP.
- https://cdn.elifesciences.org/articles/93489/elife-93489-supp2-v3.xlsx





