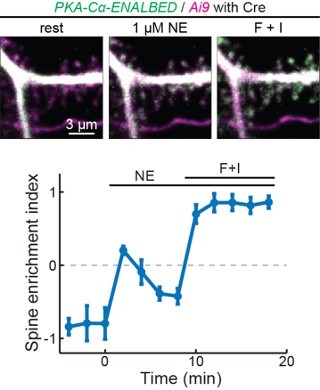
PKA regulation of neuronal function requires the dissociation of catalytic subunits from regulatory subunits
Figures
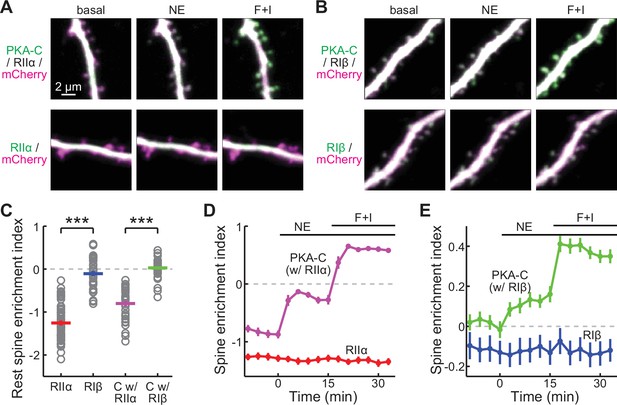
PKA-C but not PKA-R redistributes to spines upon activation.
(A, B) Representative two-photon images of PKA-C-mEGFP co-expressed with PKA-RIIα or PKA-RIβ at rest, or in the presence of norepinephrine (NE) or forskolin and IBMX (F+I). mCherry (magenta) was co-expressed to reveal the neuronal morphology. (C–E) Quantification and comparison of the spine enrichment index at the resting state (C) and upon activation (D, E). As in panel C from left to right, n (spines/neurons)=53/11, 34/7, 33/6, and 36/7. Error bars represent s.e.m.
-
Figure 1—source data 1
Numeric data for Figure 1.
- https://cdn.elifesciences.org/articles/93766/elife-93766-fig1-data1-v1.xlsx
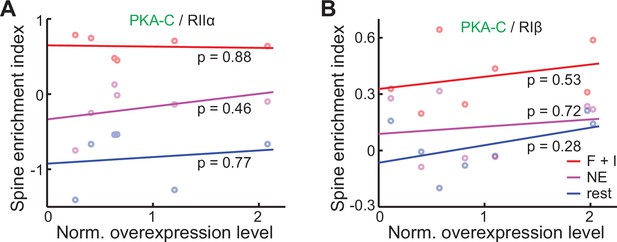
Spine enrichment indexes and the movement of PKA-C into spines upon activation are not dependent on the protein expression level.
(A–B) Averaged spine enrichment indexes within individual neurons of PKA-C co-expressed with RIIα (A) and RIβ (B), and their linear fit at rest or under the indicated stimulation.
-
Figure 1—figure supplement 1—source data 1
Numeric data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/93766/elife-93766-fig1-figsupp1-data1-v1.xlsx
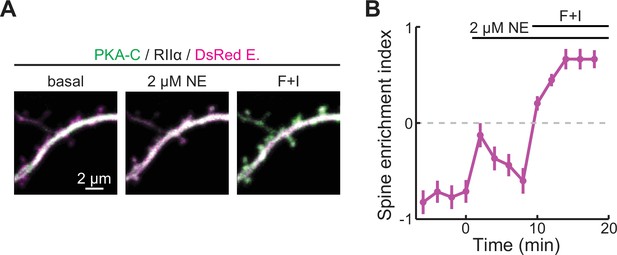
PKA-C translocation can be driven by norepinephrine at low concentrations.
(A, B) Representative two-photon images (A) and the collective trace of PKA-C-mEGFP co-expressed with PKA-RIIα at rest, or in the presence of 2 μM norepinephrine (NE) or forskolin and IBMX (F+I). DsRed Express (magenta) was co-expressed to reveal the neuronal morphology. n (spines/neurons)=16/4. Error bars represent s.e.m.
-
Figure 1—figure supplement 2—source data 1
Numeric data for Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/93766/elife-93766-fig1-figsupp2-data1-v1.xlsx
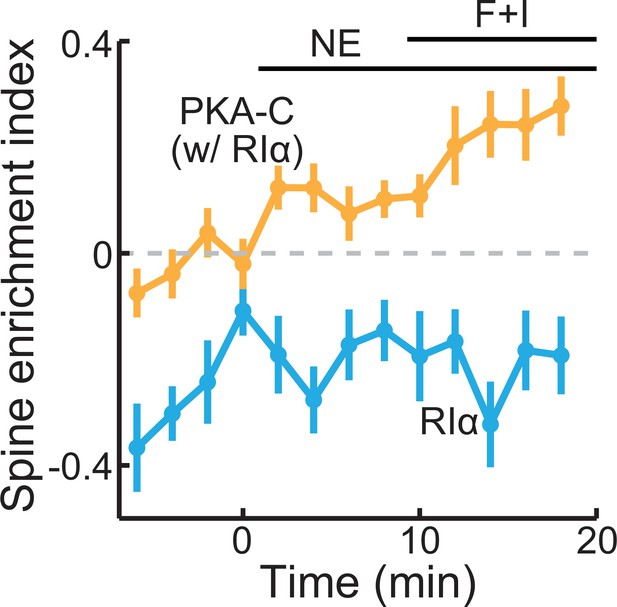
PKA-C and PKA-RIα differentially re-distributed upon activation.
The collective trace of PKA-C-mEGFP co-expressed with PKA-RIα (orange) and PKA-RIα-mEGFP (light blue) at rest, or in the presence of 10 μM norepinephrine (NE) or forskolin and IBMX (F+I). DsRed Express was co-expressed to reveal the neuronal morphology. n (spines/neurons)=20/5 for PKA-C and 16/4 for RIα. Error bars represent s.e.m.
-
Figure 1—figure supplement 3—source data 1
Numeric data for Figure 1—figure supplement 3.
- https://cdn.elifesciences.org/articles/93766/elife-93766-fig1-figsupp3-data1-v1.xlsx
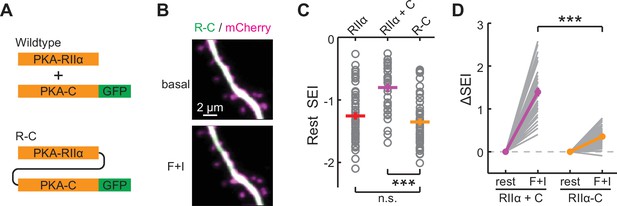
Characterization of the inseparable R-C.
(A) Schematic of wildtype PKA versus R-C. In both cases PKA-C was C-terminally tagged by mEGFP. (B–C) Representative images (B), quantifications of resting distribution (C), and the distribution change upon stimulation by forskolin and IBMX (D) of R-C compared to PKA-RIIα-mEGFP and co-expressed PKA-C-mEGFP/PKA-RIIα. RIIα and RIIα+C data are from Figure 1C. n (spines/neurons)=48/10. Error bars represent s.e.m.
-
Figure 2—source data 1
Numeric data for Figure 2.
- https://cdn.elifesciences.org/articles/93766/elife-93766-fig2-data1-v1.xlsx
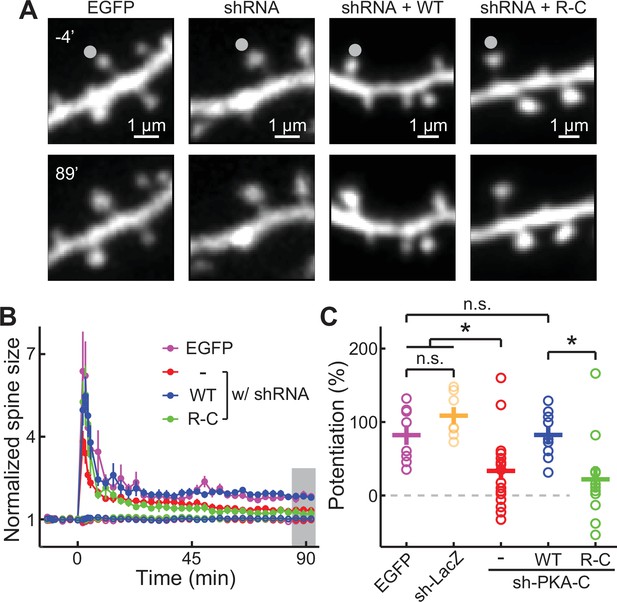
PKA regulation of synaptic plasticity cannot be sustained by an inseparable PKA variant.
(A–C) Representative image (A), time course (B), and the degree of potentiation (C) at the indicated timepoints in panel B of single-spine LTP experiments as triggered by focal glutamate uncaging at the marked spines (gray dot). In panel B, both stimulated spines (solid circles) and non-stimulated control spines (open circles) are shown. As in panel C from left to right, n (spines, each from a different neuron)=8, 7, 17, 11, 9. Error bars represent s.e.m.
-
Figure 3—source data 1
Numeric data for Figure 3.
- https://cdn.elifesciences.org/articles/93766/elife-93766-fig3-data1-v1.xlsx
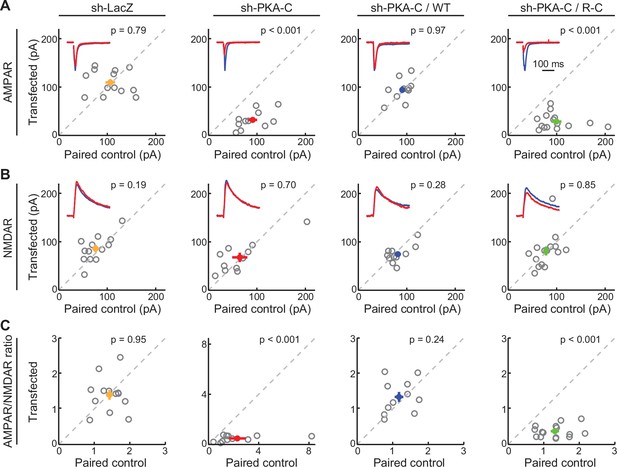
AMPA receptor-mediated synaptic transmission requires wildtype dissociable PKA.
(A–C) Representative traces (red) normalized to the paired control (blue) (insets) and scatter plots of paired AMPA (A) and NMDA (B) receptor currents and AMPA/NMDA receptor current ratios (C) from neighboring untransfected CA1 neurons and those transfected with shRNA against PKA-C and the indicated shRNA-resistant rescue constructs. Statistical p values were obtained using a sign test (MATLAB). From left to right, n (neuron pairs)=13, 11, 11, and 15. Error bars represent s.e.m.
-
Figure 4—source data 1
Numeric data for Figure 4.
- https://cdn.elifesciences.org/articles/93766/elife-93766-fig4-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Rattus norvegicus, Sprague Dawley) | Sprague Dawley rat | Charles River | Strain Code 001; RRID: RGD_734476 | |
| Recombinant DNA reagent | PKA-Cα-mEGFP (plasmid) | Addgene | # 45524; RRID: Addgene_45524 | |
| Recombinant DNA reagent | PKA-RIα-mEGFP (plasmid) | Addgene | # 45525; RRID: Addgene_45525 | |
| Recombinant DNA reagent | PKA-RIβ-mEGFP (plasmid) | Addgene | # 45526; RRID: Addgene_45526 | |
| Recombinant DNA reagent | PKA-RIIα-mEGFP (plasmid) | Addgene | # 45527; RRID: Addgene_45527 | |
| Recombinant DNA reagent | PKA-RIα (plasmid) | This paper | Figure 1—figure supplement 3 | |
| Recombinant DNA reagent | PKA-RIβ (plasmid) | This paper | Figure 1 | |
| Recombinant DNA reagent | PKA-RIIα (plasmid) | Addgene | #168492; RRID: Addgene_168492 | |
| Recombinant DNA reagent | PKA-RIIα-PKA-Cα-mEGFP (plasmid) | This paper | Figure 2 | |
| Recombinant DNA reagent | shPKA against PKA-Cα with DsRed co-expression (plasmid) | This paper | The shRNA was developed in Tillo et al., 2017; Figure 3 | |
| Recombinant DNA reagent | shPKA against LacZ with DsRed co-expression (plasmid) | This paper | Figure 3 | |
| Recombinant DNA reagent | mCherry2 (plasmid) | Addgene | #54517; RRID: Addgene_54517 | |
| Chemical compound, drug | Norepinephrine | Tocris | 5169 | |
| Chemical compound, drug | Forskolin | LC Labs | F-9926 | |
| Chemical compound, drug | IBMX | Sigma-Aldrich | I7018 | |
| Chemical compound, drug | MNI-glutamate | Tocris | 1490 | |
| Chemical compound, drug | TTX | Tocris | 1069 | |
| Chemical compound, drug | 2-Chloroadenosine | Sigma-Aldrich | C5134 | |
| Chemical compound, drug | GABAzine (SR 95531) | Tocris | 1262 | |
| Software, algorithm | MATLAB | MathWorks | RRID: SCR_001622 | |
| Software, algorithm | SI_View | Zhong, 2022 | https://github.com/HZhongLab/SI_View |