Early steps of protein disaggregation by Hsp70 chaperone and class B J-domain proteins are shaped by Hsp110
Figures
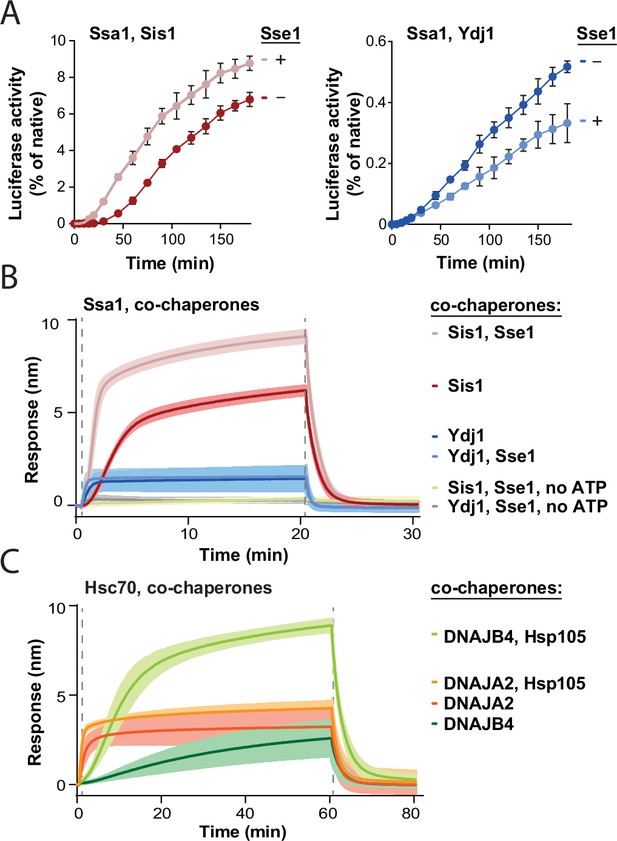
Impact of Hsp110 on protein disaggregation by Hsp70 system depends on class of J-domain protein (JDP).
(A) Refolding of aggregated luciferase by Ssa1-Sis1 (1 µM Ssa1, 1 µM Sis1)±µM Sse1 (0.1 µM) (left) or Ssa1-Ydj1 (1 µM Ssa1, 1 µM Ydj1)±Sse1 (0.1 µM) (right). Error bars show SD from three independent repeats. Luciferase activity was measured at indicated time points and normalised to the native activity. (B) Sensor-bound luciferase aggregates incubated with Ssa1-Sis1 (1 µM Ssa1, 1 µM Sis1)±Sse1 (0.1 µM) or Ssa1-Ydj1 (1 µM Ssa1, 1 µM Ydj1)±Sse1 (0.1 µM), with or without ATP, as indicated. (C) Binding of Hsc70-DNAJB4 (3 μM Hsc70, 1 μM DNAJB4) or Hsc70-DNAJA2 (3 μM Hsc70, 1 μM DNAJA2) with or without Hsp105 (0.3 μM) to the heat-aggregated luciferase immobilised on the biolayer interferometry (BLI) biosensor. (B, C) The lines represent the average of three replicates, the shades designate SD, and the dashed lines indicate the start of the chaperone binding and dissociation steps.
-
Figure 1—source data 1
Spreadsheet containing data for the graphs shown in Figure 1A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Spreadsheet containing data for the graph shown in Figure 1B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Spreadsheet containing data for the graph shown in Figure 1C.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-data3-v1.xlsx
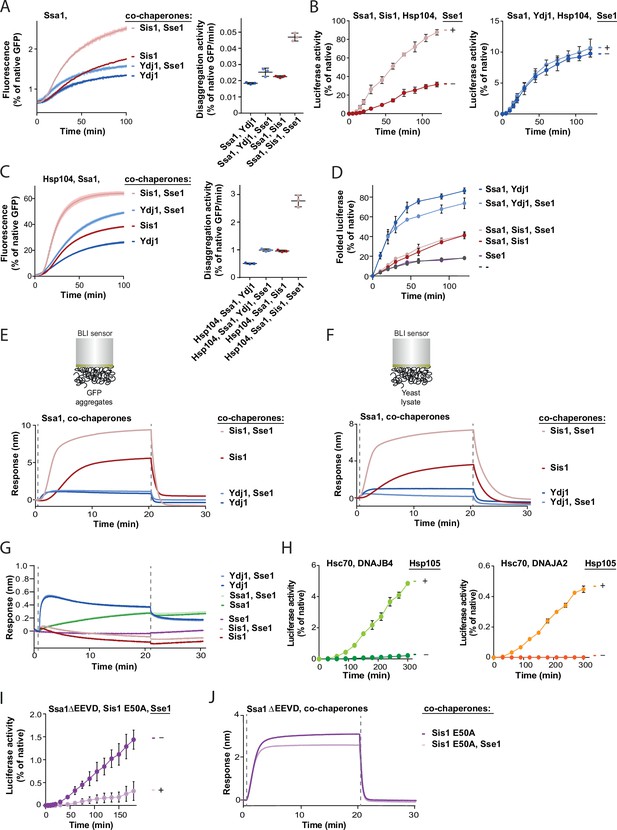
J-domain protein-specific impact of Hsp110 on protein disaggregation by Hsp70 system.
(A) Renaturation of heat-aggregated GFP by Ssa1-Sis1±Sse1 or Ssa1-Ydj1±Sse1 (1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, and 0,1 µM Sse1). (B) Refolding of aggregated luciferase by Ssa1-Sis1±Sse1 (left) or Ssa1-Ydj1±Sse1 (right) in the presence of Hsp104 (1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, 1 µM Hsp104, and 0.1 µM Sse1). (C) Renaturation of heat-aggregated GFP in the presence of Hsp104 with Ssa1-Sis1±Sse1 or Ssa1-Ydj1±Sse1 Hsp104 (1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, 1 µM Hsp104, and 0.1 µM Sse1). (D) Spontaneous folding of non-aggregated luciferase diluted from 5 M GuHCl, alone or assisted by the Hsp70 system (1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, and 0.1 µM Sse1). (E) Sensor covered with GFP aggregates or (F) yeast lysate incubated with Ssa1-Sis1±Sse1 or Ssa1-Ydj1±Sse1 (1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, and 0.1 µM Sse1). (G) Binding of the indicated combination of chaperones to luciferase aggregates immobilised on the biolayer interferometry (BLI) sensor (1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, and 0.1 µM Sse1). (H) Recovery of aggregated luciferase performed upon addition of human system comprising Hsc70-DNAJB4±Hsp105 (left) or Hsc70-DNAJA2±Hsp105 (right) (3 µM Hsc70, 1 µM DNAJB4, 1 µM DNAJA2, and 0.3 µM Hsp105). (I) Refolding of aggregated luciferase by Sis1 E50A and Ssa1 ΔEEVD±Sse1. (J) Binding of Sis1 E50A and Ssa1 ΔEEVD±Sse1 to luciferase aggregates immobilised on the BLI sensor (1 µM Ssa1 ΔEEVD, 1 µM Sis1 E50A, and 0.1 µM Sse1). Shades and error bars represent SD from three independent repeats. The BLI experiments show a result representative for at least two independent repeats. Data from all the replicates are available in Figure 1—figure supplement 1—source data 1–10.
-
Figure 1—figure supplement 1—source data 1
Spreadsheet containing data for the graphs shown in Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data1-v1.xlsx
-
Figure 1—figure supplement 1—source data 2
Spreadsheet containing data for the graphs shown in Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data2-v1.xlsx
-
Figure 1—figure supplement 1—source data 3
Spreadsheet containing data for the graphs shown in Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data3-v1.xlsx
-
Figure 1—figure supplement 1—source data 4
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 1D.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data4-v1.xlsx
-
Figure 1—figure supplement 1—source data 5
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 1E and another replicate of the experiment.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data5-v1.xlsx
-
Figure 1—figure supplement 1—source data 6
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 1F and another replicate of the experiment.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data6-v1.xlsx
-
Figure 1—figure supplement 1—source data 7
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 1G and another replicate of the experiment.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data7-v1.xlsx
-
Figure 1—figure supplement 1—source data 8
Spreadsheet containing data for the graphs shown in Figure 1—figure supplement 1H.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data8-v1.xlsx
-
Figure 1—figure supplement 1—source data 9
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 1I.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data9-v1.xlsx
-
Figure 1—figure supplement 1—source data 10
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 1J and another replicate of the experiment.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp1-data10-v1.xlsx
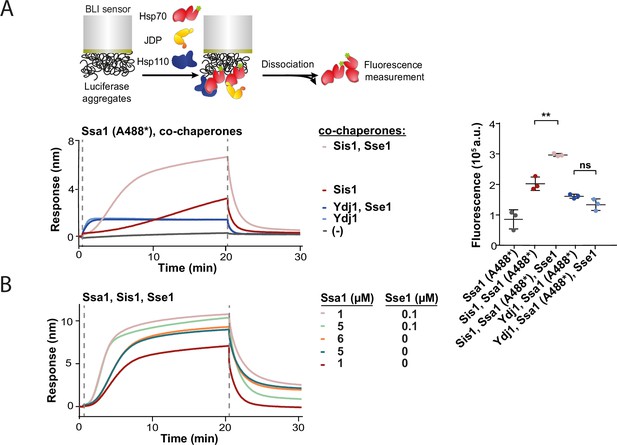
Hsp110 determines level of Hsp70 binding to aggregates.
(A) Upper panel shows the scheme of the biolayer interferometry (BLI) experiment. Sensors covered with luciferase aggregates incubated with Ssa1 labelled with Alexa 488 and Sis1 ±Sse1 or Ydj1±Sse1 (1 µM Ssa1 (A488*), 1 µM Sis1, 1 µM Ydj1, and 0.1 µM Sse1). Right panel shows fluorescence of Ssa1 (A488*) measured after the dissociation step. Two-tailed t-test: ** p<0.01, ns p>0.05 (B) Sensor-bound luciferase aggregates incubated with constant concentration of Sis1 (1 µM) and changing concentration of Ssa1 (1 µM, 5 µM, and 6 µM)±0.1 µM Sse1.
-
Figure 1—figure supplement 2—source data 1
Spreadsheet containing data for the graphs shown in Figure 1—figure supplement 2A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp2-data1-v1.xlsx
-
Figure 1—figure supplement 2—source data 2
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 2B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp2-data2-v1.xlsx
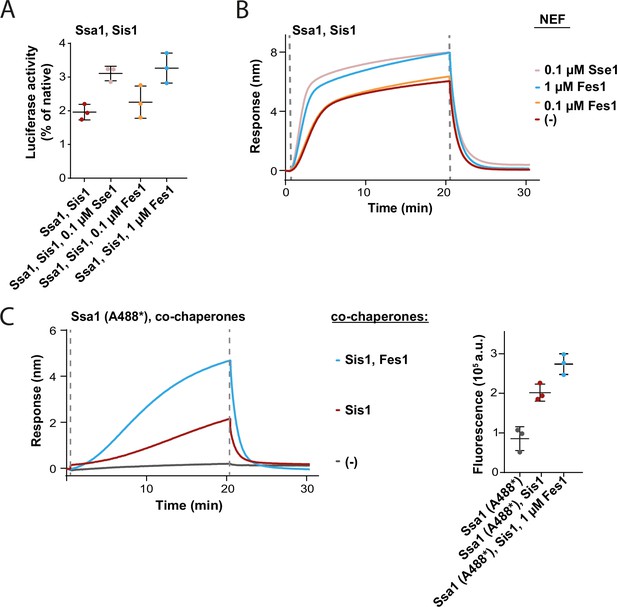
Effects of Fes1 on Hsp70 binding to protein aggregates and disaggregation.
(A) Refolding of aggregated luciferase by Ssa1-Sis1 with 0.1 µM concentration of Sse1 or increasing concentrations of Fes1 measured after 1 hr. (B) Sensor with immobilized luciferase aggregates incubated with Ssa1-Sis1 in the presence of the indicated concentrations Fes1 in comparison to Ssa1-Sis1 with 0.1 µM Sse1. (C) Sensor covered with luciferase aggregates incubated with Ssa1 labelled with Alexa 488 and Sis1 in the presence of 1 µM Fes1. Right panel shows fluorescence of Ssa1 (A488*) measured after the dissociation step. Binding kinetics together with fluorescence of Ssa1 (A488*) and Sis-Ssa1 (A488*) was adapted from Figure 1G. Error bars show SD from three independent repeats. Biolayer interferometry (BLI) curves show a result representative for at least two independent repeats. Data from all the replicates are available in Figure 1—figure supplement 3—source data 1–3.
-
Figure 1—figure supplement 3—source data 1
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 3A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp3-data1-v1.xlsx
-
Figure 1—figure supplement 3—source data 2
Spreadsheet containing data for the graph shown in Figure 1—figure supplement 3B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp3-data2-v1.xlsx
-
Figure 1—figure supplement 3—source data 3
Spreadsheet containing data for the graphs shown in Figure 1—figure supplement 3C.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig1-figsupp3-data3-v1.xlsx
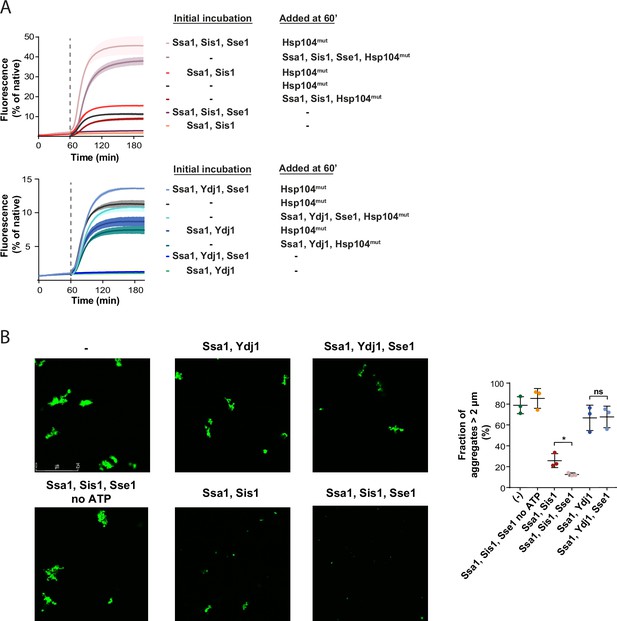
Sse1 promotes modification of aggregates by the Hsp70 system.
(A) Initial incubation of heat-aggregated GFP aggregates with the Hsp70 system, followed by the addition of the Hsp104 D484K F508A variant (0.15 µM). Recovery was initiated by the addition of the mix of indicated chaperones: 1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, or 0.1 µM Sse1. Dashed lines indicate the beginning of the incubation with the Hsp104 variant. Curves show average values and shades indicate SD from three replicates. (B) Fluorescence microscopy images of luc-GFP monitored upon addition of Ssa1-Sis1±Sse1 or Ssa1-Ydj1±Sse1. Chaperones were used at 1 µM concentration, except for 0.1 µM Sse1. Left panels show controls of the luciferase-GFP aggregates alone and upon the addition of the Hsp70 system without ATP. Quantification of the fraction of aggregates >2 µm is from three independent replicates. Two-tailed t test was performed: *p<0.05, ns: not significant.
-
Figure 2—source data 1
Spreadsheet containing data for the graph shown in Figure 2A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Spreadsheet containing data for the graph shown in Figure 2B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Uncropped microscopy images presented in Figure 2B and replicates for the calculations in Figure 2—source data 2.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig2-data3-v1.zip
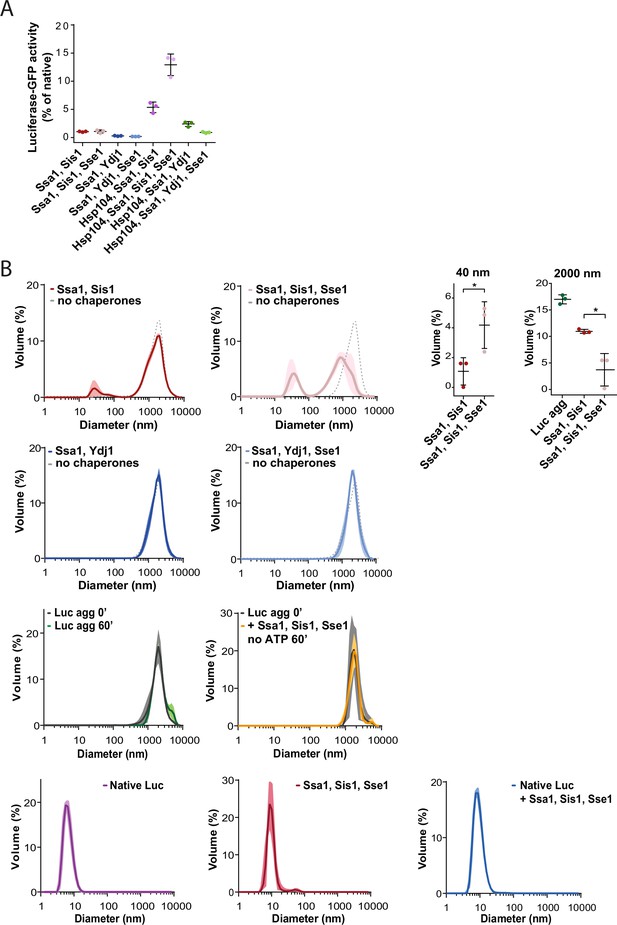
Hsp110 impact on aggregate modification by Hsp70 system.
(A) Recovery of FLUC-EGFP aggregates (0.3 µM) by Hsp70 system with indicated combination of Ssa1 (1 µM), Sis1 (1 µM), Ydj1 (1 µM), Hsp104 (1 µM), or Sse1 (0.1 µM). luc-GFP activity was measured after 1 hr incubation and normalised to the native activity. Error bars indicate SD from three repeats. (B) Size distribution measured with dynamic light scattering. Diameter of luciferase aggregates incubated alone or with Ssa1-Sis1±Sse1 or Ssa1-Ydj1±Sse1, as indicated in the figure, with ATP, unless stated otherwise, was measured after 1 hr incubation. Proteins were used at the same concentrations as in A. Additionally, measurements with native luciferase, chaperones, or chaperones with native luciferase were performed (lower panels). Lines are the average of three replicates while the shades indicate standard deviation. Dashed lines designate the size distribution of the luciferase aggregates prior to the addition of chaperones. Upper right panel shows the height of the peak at 40 nm and 2000 nm for Ssa1-Sis1 with or without 0.1 µM Sse1, analysed with the two-tailed t test: *p<0,05, **p<0,01 using the GraphPad Prism software.
-
Figure 2—figure supplement 1—source data 1
Spreadsheet containing data for the graph shown in Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig2-figsupp1-data1-v1.xlsx
-
Figure 2—figure supplement 1—source data 2
Spreadsheet containing data for the graphs shown in Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig2-figsupp1-data2-v1.xlsx
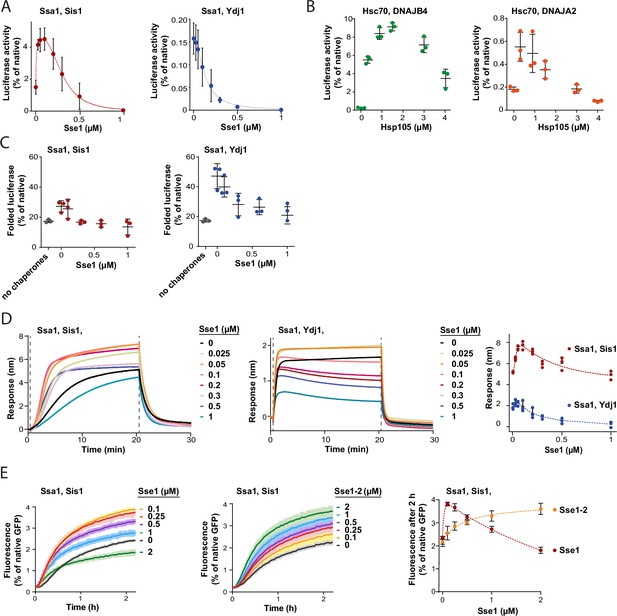
Susceptibility of Hsp70 to Hsp110 depends on J-domain protein (JDP) class and phase of disaggregation.
(A) Titration of Sse1 in the refolding of aggregated luciferase by Ssa1-Sis1 (1 µM Ssa1, 1 µM Sis1) (red) or Ssa1-Ydj1 (1 µM Ssa1, 1 µM Ydj1) (blue). Activity of luciferase was measured after 1 hr and normalised to that of the native protein. (B) Incubation of Hsc70-DNAJB4 (3 µM Hsc70, 1 µM DNAJB4) (green) or Hsc70-DNAJA2 (3 µM Hsc70, 1 µM DNAJA2) (orange) with luciferase aggregates at increasing concentrations of Hsp105. Luciferase activity was measured after 4 hr and normalised to the activity of the native protein. (C) Folding of non-aggregated luciferase diluted from 5 M GuHCl (grey), spontaneous or mediated by the Hsp70 system comprising Ssa1-Sis1 (1 µM Ssa1, 1 µM Sis1) (red) or Ydj1-Ssa1 (1 µM Ssa1, 1 µM Ydj1) (blue) with increasing concentrations of Sse1. Activity of luciferase was measured after 2 hr and normalised to the native protein. (D) Binding of Ssa1-Sis1 (1 µM Ssa1, 1 µM Sis1) or Ssa1-Ydj1 Ssa1 (1 µM Ssa1, 1 µM Ydj1) in the presence of Sse1 at the indicated concentrations to the sensor covered with luciferase aggregates. Right panel shows a plot of the binding signal prior to the dissociation step. (E) Renaturation of heat-aggregated GFP by Ssa1-Sis1 (1 µM Ssa1, 1 µM Sis1) in the presence of Sse1 or Sse1-2 at the indicated concentrations. Right panel shows the plot of the recovered GFP activity after 2 hr of incubation with the Hsp70 system in the presence of Sse1 (orange) or Sse1-2 (red). (D, E) Dashed lines show the fitting of the [Agonist] vs response model to the data from the stimulation and inhibition phases separately using the GraphPad Prism software. (A–E) Error bars and shades indicate SD from three repeats.
-
Figure 3—source data 1
Spreadsheet containing data and model fitting parameters for the graphs shown in Figure 3A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Spreadsheet containing data for the graphs shown in Figure 3B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-data2-v1.xlsx
-
Figure 3—source data 3
Spreadsheet containing data for the graphs shown in Figure 3C.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Spreadsheet containing data for the graphs shown in the left and middle panels of Figure 3D.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-data4-v1.xlsx
-
Figure 3—source data 5
Spreadsheet containing data for the graph shown in the right panel of Figure 3D.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-data5-v1.xlsx
-
Figure 3—source data 6
Spreadsheet containing data for the graphs shown in the left and middle panels of Figure 3E.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-data6-v1.xlsx
-
Figure 3—source data 7
Spreadsheet containing data and model fitting parameters for the graph shown in the right panel of Figure 3E.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-data7-v1.xlsx
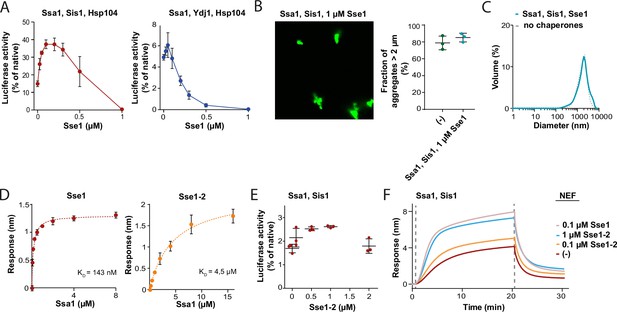
Concentration-dependent effects of Hsp110 on aggregate binding and disaggregtion by Hsp70.
(A) Titration of Sse1 in the refolding of aggregated luciferase by Ssa1-Sis1 (red) or Ssa1-Ydj1 (blue) with Hsp104 (1 µM Ssa1, 1 µM Sis1, 1 µM Ydj1, 1 µM Hsp104, and indicated concentrations of Sse1). Activity of luciferase was measured after 1 hr and normalised to the native activity. Shown is average and SD from three repeats. (B) Fluorescence microscopy images of luciferase-GFP aggregates incubated with Ssa1 (1 µM) and Sis1 (1 µM) in the presence of 1 µM Sse1. Scale is as in Figure 2B. Error bars show SD from three repeats. Quantification of the fraction of aggregates >2 µm is from three independent replicates. Data for aggregates alone are from Figure 2B. (C) Size distribution of luciferase aggregates incubated with Ssa1 (1 µM) and Sis1 (1 µM) in the presence of 1 µM Sse1 measured by dynamic light scattering. (D) Dissociation constant is determined based on the level of Ssa1 binding to Sse1 or Sse1-2. Sse1 or Sse1-2 was immobilised on the biolayer interferometry (BLI) sensor through His6-SUMO tag and Ssa1 was used at concentrations: 16,000, 8000, 4000, 2000, 1000, 500, 250, 125, 63, and 32 nM. Points indicate mean with SD from three independent experiments. The One site-specific binding model was fitted to the data with the GraphPad Prism software. (E) Titration of Sse1-2 in the refolding of luciferase aggregates assay with Ssa1-Sis1 (1 µM Ssa1, 1 µM Sis1). (F) Luciferase aggregates immobilised on the BLI sensor were incubated with Ssa1-Sis1 with either Sse1 or Sse1-2 at the indicated concentrations (1 µM Ssa1, 1 µM Sis1).
-
Figure 3—figure supplement 1—source data 1
Spreadsheet containing data for the graphs shown in Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-figsupp1-data1-v1.xlsx
-
Figure 3—figure supplement 1—source data 2
Spreadsheet containing data for the graph shown in Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-figsupp1-data2-v1.xlsx
-
Figure 3—figure supplement 1—source data 3
Uncropped microscopy images presented in Figure 3—figure supplement 1B and replicates for the calculations in Figure 3—figure supplement 1—source data 2.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-figsupp1-data3-v1.zip
-
Figure 3—figure supplement 1—source data 4
Spreadsheet containing data for the graph shown in Figure 3C.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-figsupp1-data4-v1.xlsx
-
Figure 3—figure supplement 1—source data 5
Spreadsheet containing data for the graphs shown in Figure 3D.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-figsupp1-data5-v1.xlsx
-
Figure 3—figure supplement 1—source data 6
Spreadsheet containing data for the graph shown in Figure 3E.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-figsupp1-data6-v1.xlsx
-
Figure 3—figure supplement 1—source data 7
Spreadsheet containing data for the graph shown in Figure 3F.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig3-figsupp1-data7-v1.xlsx
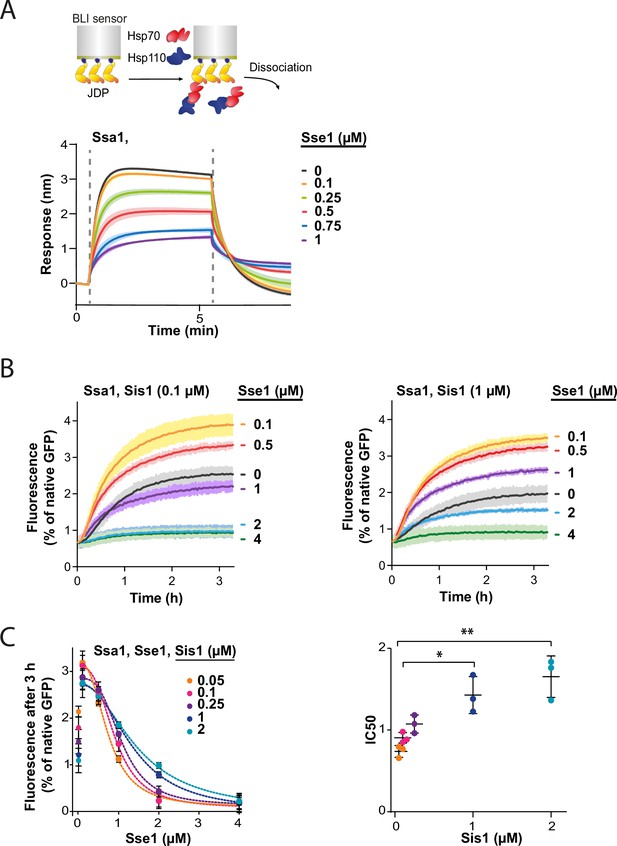
Hsp110 and class B J-domain protein (JDP) show apparent competition for Hsp70.
(A) Upper panel shows the scheme of the biolayer interferometry (BLI) experiment. Binding of Ssa1 (1 µM) in the presence of increasing concentrations of Sse1 to Sis1 immobilised on the BLI sensor through the His6-SUMO tag. Dashed lines indicate the moment of addition of chaperones to the sensor-bound Sis1 and the dissociation step. (B) Renaturation of heat-aggregated GFP by Ssa1-Sis1 (1 µM Ssa1, 0.1 µM, or 1 µM Sis1) at increasing concentrations of Sse1. (C) Plot of GFP activity after 3 hr recovered from aggregates by Ssa1 at different concentrations of Sis1 and Sse1 (left). IC50 was determined by fitting the [Inhibitor] versus response model to the data from three experiments using the GraphPad Prism software (dashed lines). Two-tailed t test: *p<0.05, **p<0.01. Lines are the average of three replicates and the error bars and shades designate standard deviation.
-
Figure 4—source data 1
Spreadsheet containing data for the graph shown in Figure 4A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Spreadsheet containing data for the graphs shown in Figure 4B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Spreadsheet containing data and model fitting parameters for the graphs shown in Figure 4C.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-data3-v1.xlsx
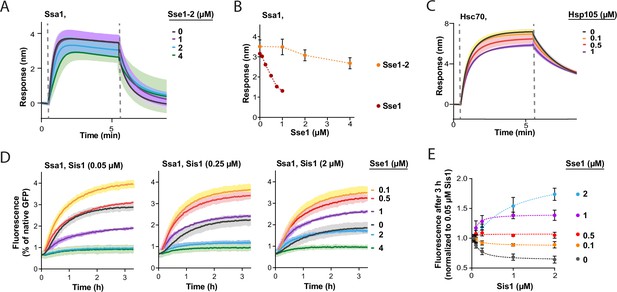
Competition between Hsp110 and JDP co-chaperones.
(A) Titration of Sse1-2 in the presence of 1 µM Ssa1 in the binding to the immobilised Sis1 on the biolayer interferometry (BLI) sensor. (B) Comparison of the binding signal prior to the dissociation of Ssa1 in the presence of the indicated concentrations of Sse1 or Sse1-2. The binding signal of Ssa1-Sse1 wild type (WT) was adapted from Figure 4A. (C) Incubation of His6-SUMO-DNAJB4 immobilised on the BLI sensor with Hsc70 and the indicated concentrations of Hsp105. (D) Titration of Sse1 in the GFP reactivation assay with Ssa1 (1 µM) and Sis1 at different concentrations: 0.05 µM, 0.25 µM, 2 µM. (E) Comparison of GFP fluorescence monitored after 3 hr of incubation with Ssa1, Sis1, and Sse1. The values were adapted from (D) and for 0.1 µM and 1 µM Sis1 they were adapted from Figure 4B. Dashed lines show fitting of the [Agonist] versus response model to the data from three experiments using the GraphPad Prism software. Lines and points represent average values and error bars and shades indicate SD from three independent replicates.
-
Figure 4—figure supplement 1—source data 1
Spreadsheet containing data for the graph shown in Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-figsupp1-data1-v1.xlsx
-
Figure 4—figure supplement 1—source data 2
Spreadsheet containing data for the graph shown in Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-figsupp1-data2-v1.xlsx
-
Figure 4—figure supplement 1—source data 3
Spreadsheet containing data for the graph shown in Figure 4—figure supplement 1C.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-figsupp1-data3-v1.xlsx
-
Figure 4—figure supplement 1—source data 4
Spreadsheet containing data for the graphs shown in Figure 4—figure supplement 1D.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-figsupp1-data4-v1.xlsx
-
Figure 4—figure supplement 1—source data 5
Spreadsheet containing data for the graph and model fitting parameters shown in Figure 4—figure supplement 1E.
- https://cdn.elifesciences.org/articles/94795/elife-94795-fig4-figsupp1-data5-v1.xlsx
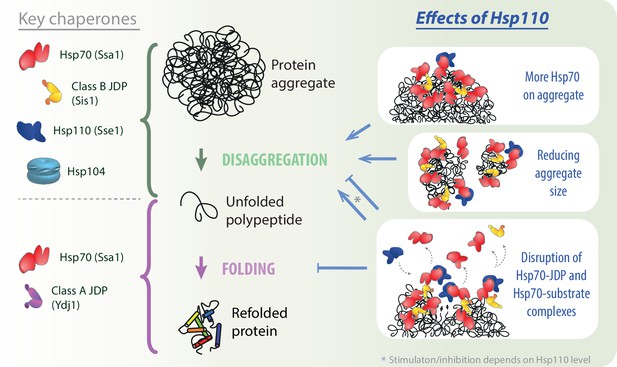
Impact of Hsp110 on Hsp70-dependent disaggregation.
Hsp110 stimulates aggregate disassembly by Hsp70 with class B J-domain proteins (JDPs) and Hsp104 (green arrow), contrary to the stage of final folding of the solubilised polypeptide (light purple arrow), at which Hsp70 with class A JDP are most effective. Hsp110 improves the disaggregation by increasing Hsp70 binding to the aggregate (upper right panel) and mediates aggregate remodelling into smaller assemblies (middle right panel). By facilitating partial Hsp70 dissociation from the substrate and/or JDP, at the optimal, sub-stoichiometric concentration, Hsp110 might gradually uncover new Hsp70-binding sites (lower right panel, small black arrows), potentially leading to the more abundant and effective Hsp70 recruitment to the aggregate. At higher Hsp110 levels, the destabilisation of Hsp70 interactions with protein substrates and JDPs leads to the inhibition across all stages of protein disaggregation (lower right panel).

Chaperone-assisted folding of luciferase after denaturation at 5 M or 6 M GdnHCl.
Luciferase was denatured in 5 M or 6 M GdnHCl according to the protocol in the Materials and Methods section. Luminescence was monitored alone or after incubation with Luminescence was monitored alone or after incubation with Ssa1-Sis1 or Ssa1-Ydj1. Chaperones were used at 1 µM concentration. Luciferase activity was measured after 2 hours and normalized to the activity of the native protein. Error bars indicate SD from three repeats.

Disaggregaton of heat-aggregated luciferase – impact of sHsps.
Luciferase (2 μM) was denatured with (blue) or without (red) Hsp26 (20 μM) at 45 ̊C for 15 min in the buffer A (Materials and Methods). Upon 100-fold dilution with the buffer A, supplemented wih 5 mM ATP, 2 mM DTT, 1.2 μM creatine kinase, 20 mM creatine phosphate, chaperones indicated in the legend were added to the final concentration of 1 μM, except for Sse1, concentration of which was 0.1 μM. Shown is luciferase activity measured after 1 h of incubation at 25 °C, normalized to the activity of native luciferase.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) CodonPlus | Agilent | Cat # 230250; RRID:SCR_013575 | Genotype: E. coli B F- ompT hsdS(rB- mB-) dcm +Tetr gal endA Hte [argU proL Camr] |
| Strain, strain background (E. coli) | RosettaBL21 (DE3) | Novagen | Cat # 70954; RRID:SCR_008441 | Genotype: E. coli F- ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE (CamR) |
| Strain, strain background (Saccharomyces cerevisiae) | W303 | Laboratory collection | Genotype: MATa/MATα {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15} [phi+] | |
| Peptide, recombinant protein | Ssa1 | DOI:10.1073/pnas.0804187105 | Uniprot ID: P10591 | Expressed from pCA533-His6-SUMO-SSA1 plasmid, KanR, T7 |
| Peptide, recombinant protein | Ssa1 ∆EEVD | DOI:10.1016/j.jmb.2015.02.007 | Expressed from pCA533-His6-SUMO-SSA1 ∆EEVD plasmid, KanR, T7 | |
| Peptide, recombinant protein | Sse1 | DOI:10.1073/pnas.0804187105 | Uniprot ID: P32589 | Expressed from pCA534-SSE1 plasmid KanR, T7 |
| Peptide, recombinant protein | Sse1-2 (N572Y N575A) | This study | Variant generated based on DOI:10.1016/j.jmb.2010.07.004, expressed from pCA534-SSE1-2 plasmid KanR, T7 | |
| Peptide, recombinant protein | Fes1 | DOI:10.1073/pnas.0804187105 | Uniprot ID: P38260 | Expressed from pCA707-FES1 plasmid KanR, T7 |
| Peptide, recombinant protein | Ydj1 | DOI:10.1074/jbc.M112.387589 | Uniprot ID: P25491 | Expressed from pET21a-YDJ1 plasmid AmpR, T7 |
| Peptide, recombinant protein | Sis1 | DOI:11810.1073/pnas.2108163118 | Uniprot ID: P25294 | Expressed from pPROEX-TEV-SIS1 plasmid AmpR, T7 |
| Peptide, recombinant protein | Sis1 E50A | DOI:10.1016/j.jmb.2015.02.007 | Expressed from pPROEX-TEV-SIS1 E50A plasmid AmpR, trc | |
| Peptide, recombinant protein | Hsp104 | DOI:10.1074/jbc.M112.387589 | Uniprot ID: P31539 | Expressed from pET5a-HSP104 plasmid AmpR, T7 |
| Peptide, recombinant protein | Hsp104 D484K F508A | DOI:10.1016/j.jmb.2019.04.014 | Expressed from pET5a-HSP104 D484K F508A plasmid AmpR, T7 | |
| Peptide, recombinant protein | Hsc70 | DOI:10.1038/nature14884 | Uniprot ID: P11142 | Expressed from pPROEX-His-TEV-HSC70 plasmid AmpR, trc |
| Peptide, recombinant protein | DNAJA2 | DOI:10.1038/nature14884 | Uniprot ID: O60884 | Expressed from pPROEX-His-TEV-DNAJA2 plasmid AmpR, trc |
| Peptide, recombinant protein | DNAJB1 | DOI:10.1038/nature14884 | Uniprot ID: P25685 | Expressed from pPROEX-His-TEV-DNAJB1 plasmid AmpR, trc |
| Peptide, recombinant protein | Hsp105 | DOI:10.1038/nature14884 | Uniprot ID: Q92598 | Expressed from pPROEX-His-TEV-HSP105 plasmid AmpR, trc |
| Peptide, recombinant protein | GFP | DOI:10.1074/jbc.M402405200 | Expressed from pGFPuv plasmid (TaKaRa, RRID:SCR_003960) | |
| Peptide, recombinant protein | Fluc-EGFP | This study | EGFP fusion with firefly luciferase, expressed from pET22b-Fluc-GFP plasmid AmpR, T7 | |
| Peptide, recombinant protein | His-tagged EGFP | This study | ||
| Peptide, recombinant protein | OuantiLum Recombinant Luciferase | Promega | Cat # E1701; RRID:SCR_006724 | |
| Peptide, recombinant protein | His-tagged Luciferase | DOI:10.1371/journal.pgen.1008479 | ||
| Peptide, recombinant protein | Creatine Kinase | Roche | Cat # 10127566001; RRID:SCR_001326 | |
| Commercial assay or kit | Luciferase Assay System | Promega | Cat # E151A; RRID:SCR_006724 | |
| Chemical compound, drug | Alexa Fluor 488 C5 Maleimide | Invitrogen | Cat # A10254; RRID:SCR_013378 | |
| Commercial assay or kit | QuikChange Site-Directed Mutagenesis Kit | Agilent | Cat # 200513; RRID:SCR_013575 | |
| Other | Ni-NTA BLI sensors | Sartorius | Cat # 18-5101; RRID:SCR_003935 |





