The role of RNA in the maintenance of chromatin domains as revealed by antibody-mediated proximity labelling coupled to mass spectrometry
Figures
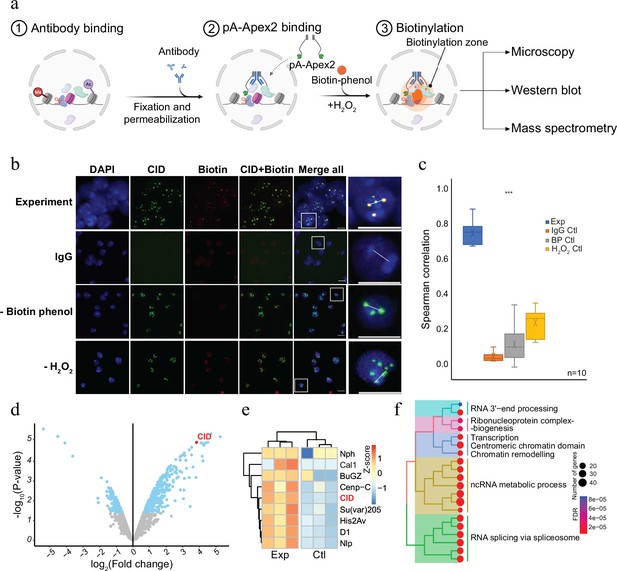
Antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS) to study the proteomic composition of the Drosophila centromere in a genetically unperturbed cell line.
(a) Schematic of AMPL-MS. Isolated nuclei were fixed, permeabilized, and incubated with specific antibodies. Recombinant pA-Apex2 enzyme binds to the antibody and biotinylates associated proximal proteins upon the addition of H2O2 and biotin-phenol. Proximal protein biotinylation is visualized by microscopy, western blot and proteins were identified by mass spectrometry. Created with BioRender.com. The schematic (b) immunofluorescence microscopy of centromeres using a Cenp-A (CID) antibody (in green) and the corresponding proximity proteome after biotinylation by pA-Apex2 (in red). Nuclear DNA was stained by 4′,6-diamidino-2-phenylindole (DAPI, in blue). IgG was used as antibody control. Scale bars represent 10 µm (large panel) or 5 µm (small panel). (c) Distribution of pair-wise Spearman correlations for quantifying the relationship between CID and biotinylation. Images of 10 cells from three independent experiments were used. Statistical significance is based on Wilcoxon rank sum test (***p-value <0.001). (d) Volcano plot of purified biotinylated proteins identified by mass spectrometry. The bait protein is highlighted in red. The x-axis represents the log2 fold change and the y-axis represents −log10 p-value comparing three IgG replicates with three CID antibody replicates (paired). The significantly enriched proteins (abs(LFC) >1 and padj ≤ 0.01) are highlighted in blue. (e) Heatmap showing the enrichment of known centromeric proteins. The heatmap was plotted using scaled log2 raw intensities. Each column represents values obtained from three independent biological replicates. (f) Over representation analysis showing top 20 biological processes (BP) using significant proteins from (d). Unsupervised clustering was performed for the gene ontology (GO) terms. The colour gradation form blue to red represents FDR (false discovery rate) and dot size represents the number of proteins found enriched in the named pathway (count).
© 2024, BioRender Inc. Figure 1a was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
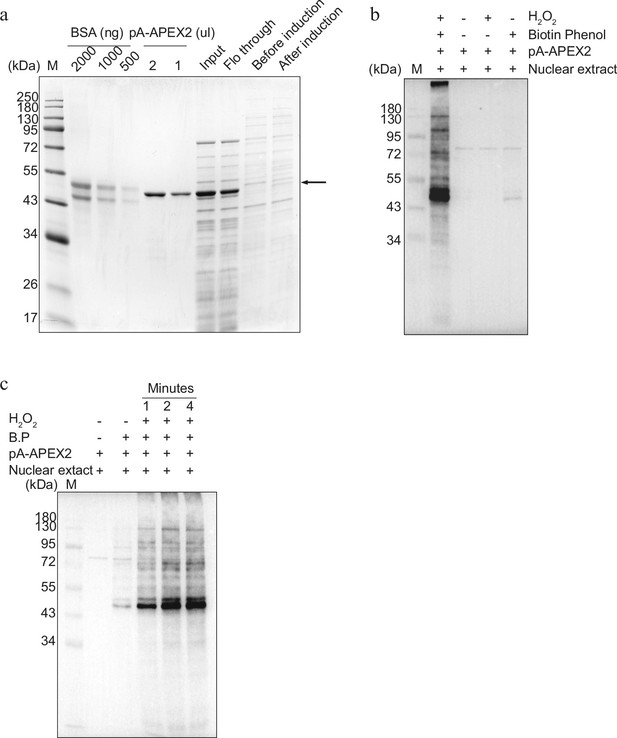
Establishment of protein A–Apex2- (pA-Apex2) mediated biotinylation for antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS) (related to Figure 1).
(a) Coomassie stained gel showing expression, purification, and quantification of pA-Apex2 enzyme. (b) In vitro activity assay of the purified pA-Apex2 enzyme. pA-Apex2 was incubated with Drosophila nuclear extract in presence of biotin-phenol and activated with H2O2, along with three different controls as marked on top of the western blot image (for details please refer to the Material and methods section). (c) Time course for the activity of pA-Apex2 as above (b).
-
Figure 1—figure supplement 1—source data 1
Raw image files of the Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/95718/elife-95718-fig1-figsupp1-data1-v2.zip
-
Figure 1—figure supplement 1—source data 2
Raw image files of the Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/95718/elife-95718-fig1-figsupp1-data2-v2.zip
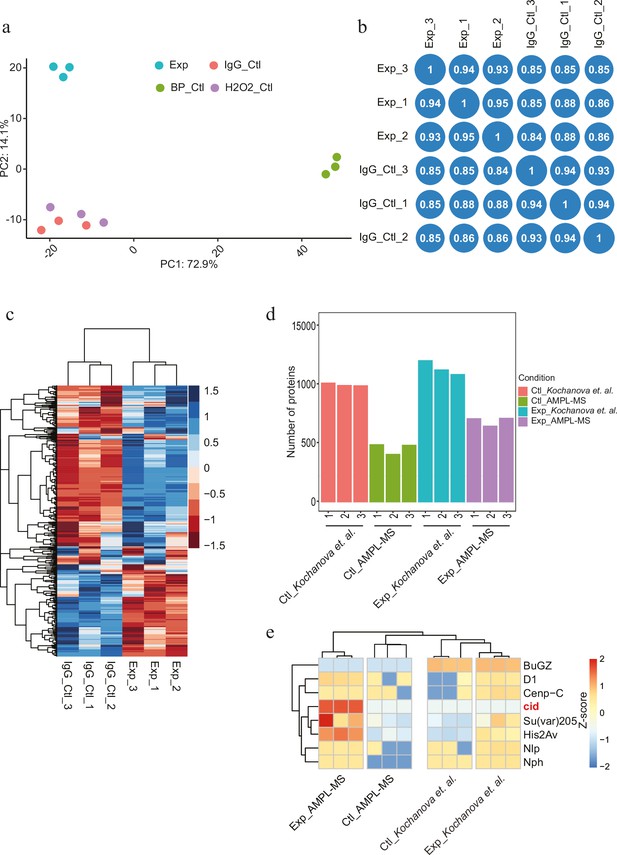
Antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS) for Drosophila centromere (related to Figure 1).
(a) Principal component analysis (PCA) between AMPL-MS of CID experiment, IgG experiment (antibody control), CID-biotin-phenol control, and CID H2O2 control. (b) Correlation plot showing reproducibility of three AMPL-MS replicates in terms of log2 FC correlation; Pearson’s correlation (r) is reported in each block. (c) Heatmap of differential abundance of proteins from CID AMPL-MS and control antibody IgG AMPL-MS. Unsupervised clustering was performed based on protein abundance. The rows represent protein and the column represents different sample. (d) A bar plot showing the number of proteins identified in CID AMPL-MS in comparison to exogenous plasmid expression-based CID_Apex2 proximity proteomics experiment (Kochanova et al., 2020). (e) Heatmap showing a comparison for enrichment of known centromeric proteins across experiment. The heatmap was plotted using scaled log2 raw intensities. Each column represents values obtained from three independent biological replicates.
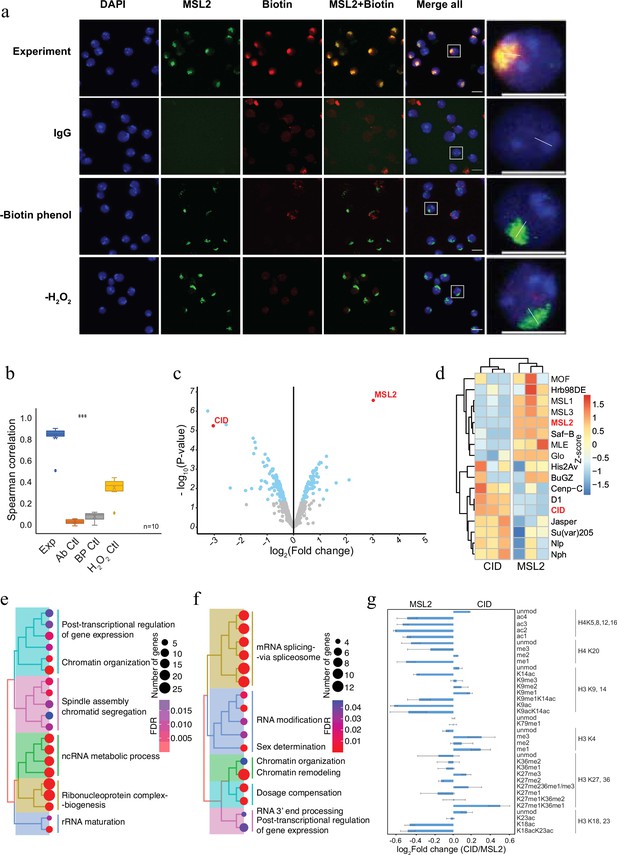
Antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS) can efficiently identify protein associated with different chromatin domains.
(a) Immunofluorescence of the X chromosome bound by MSL2 antibody (in green) and biotinylated proteins after biotinylation by pA-Apex2 (in red). Nuclei were stained by DAPI (in blue) and IgG was used as antibody control. Scale bars represent 10 µm (large panel) or 5 µm (small panel). (b) Ten cells from three independent experiments were used to quantifying the relationship between MSL-2 and biotinylation. Images of 10 cells from three independent experiments were used. Wilcoxon rank sum test is used for comparison (***p-value <0.001). (c) Volcano plot of biotinylated proteins identified by mass spectrometry. The bait proteins are highlighted in red. The x-axis represents the log2 fold change and the y-axis represents −log10 p-value comparing three CID replicates with three MSL-2 antibody replicates (paired). The significantly enriched proteins (Log2fold change(LFC) >1 and padj ≤ 0.01) are highlighted in blue. (d) Heatmap displaying the enrichment of known centromeric and X-chromosome-associated proteins. The heatmap was plotted using scaled log2 raw intensities. Each column represents values obtained from three independent biological replicates. Over representation analysis showing top 20 biological process (BP) for significantly enriched proteins from (c) associated with either centromere (e) or the X chromosome (f). The colour gradation form blue to red represents FDR (false discovery rate) and dot size represents the number of proteins found enriched in the named pathway (count). (g) Relative quantification of histone modifications associated with the X chromosome territory (left) and the centromere (right).
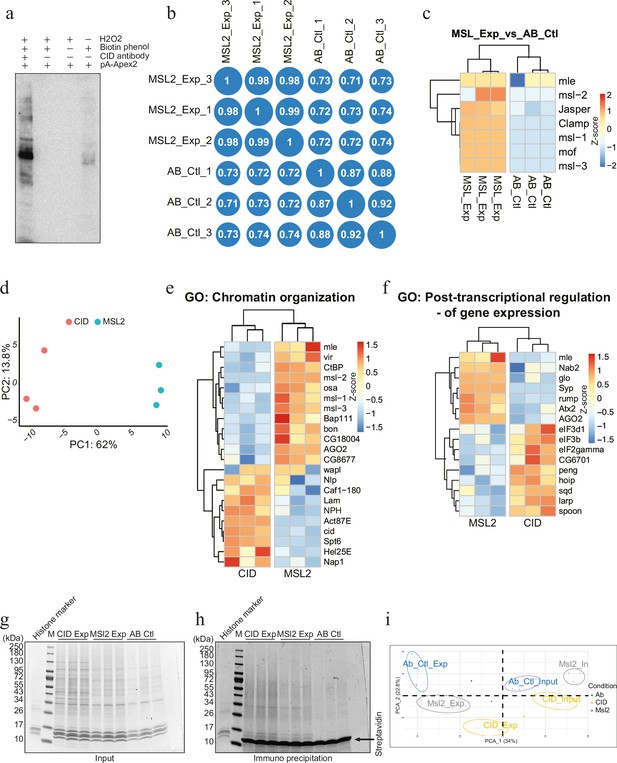
Proximity proteomics for Drosophila X chromosome and its comparison to the centromeric chromatin domain (related to Figure 2).
(a) Immunoprecipitation of biotinylated proteins after MSL2 antibody-mediated proximity biotinylation assay. (b) Correlation plot showing reproducibility of MSL2 antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS) and corresponding antibody control; Pearson’s correlation (r) is reported in each block. (c) Heatmap showing enrichment of member of DCC (dosage compensation complex); proteins known to associated with hyperactive X chromosome. The heatmap was plotted using scaled log2 raw intensities. Each column represents values obtained from three independent biological replicates. (d) Principal component analysis (PCA) between AMPL-MS of CID and MSL2. (e) Heatmap showing enrichment of protein associated with gene ontology (GO) term: ‘Chromatin organization’. The heatmap was plotted using scaled log2 raw intensities. Each column represents values obtained from three independent biological replicates. (f) Heatmap showing enrichment of protein associated with GO term: ‘Post-transcriptional regulation of gene expression’. The heatmap was plotted as above. (g) Coomassie stained gel showing the input sample used for immunoprecipitation of histones associated with CID and MSL2 AMPL-MS, used for histone modification analysis. (h) Coomassie stained gel after immunoprecipitation for biotin which subsequently used for histone analysis. (i) PCA for histone modification for samples in (h).
-
Figure 2—figure supplement 1—source data 1
Raw image files of the Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/95718/elife-95718-fig2-figsupp1-data1-v2.zip
-
Figure 2—figure supplement 1—source data 2
Labelled raw image files of the Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/95718/elife-95718-fig2-figsupp1-data2-v2.zip
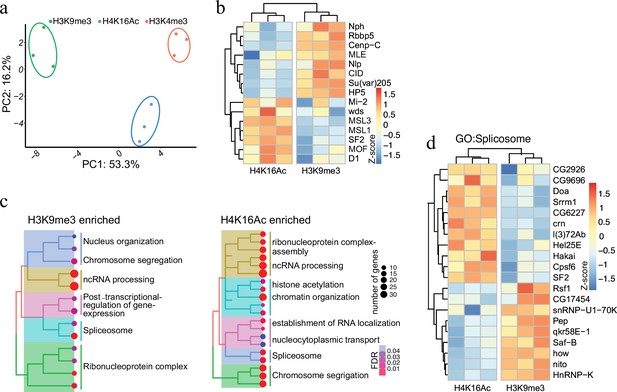
Protein associated with different and dynamic histone modifications can be effectively identified by antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS).
(a) Principal component analysis (PCA) based on the proteomic composition for three different histone marks: H3K9me3, H4K16ac, and H3K4me3 antibodies. (b) Heatmap displaying the enrichment of known H3K9me3 and H4K16ac histone modification associated proteins. The heatmap was plotted using scaled log2 raw intensities. Each column represents values obtained from three independent biological replicates. (c) Over representation analysis showing top 20 biological process (BP) for the significantly enriched protein associated with either H3K9me3 or H4K16ac histone modifications. The colour gradation form blue to red represents FDR (false discovery rate) and dot size represents the number of proteins found enriched in the named pathway (count). (d) Heatmap for protein categorized under the gene ontology (GO) term ‘Spliceosome’, a common GO term found for proteins associated with H3K9me3 and H4K16ac histone modification. The heatmap was plotted using scaled log2 raw intensities. Each column represents values obtained from three independent biological replicates.
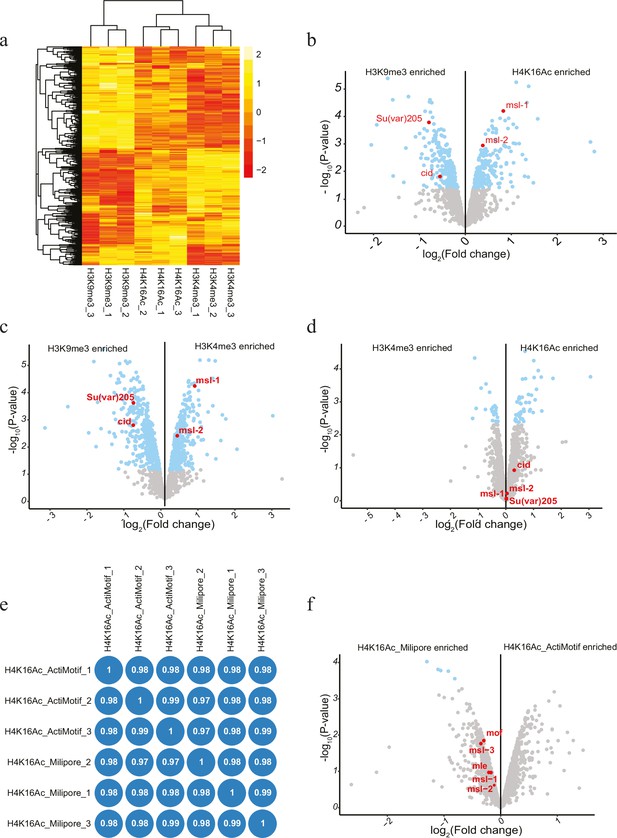
Antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS) for histone modification (related to Figure 3).
(a) Heatmap of differential abundance of proteins from AMPL-MS of H3K9me3, H4K16ac, and H3K4me3. Unsupervised clustering was performed based on protein abundance. The rows represent protein and the column represents different sample. (b) Volcano plot of biotinylated proteins associated with H3K9me3 and H4K16Ac histone modification, as identified by mass spectrometry. Two of the known associated proteins are highlighted in red. The x-axis represents the log2 fold change and the y-axis represents −log10 p-value comparing three H3K9me3 AMPL-MS replicates with three H3K4me3 AMPL-MS replicates (paired). The significantly enriched proteins (LFC >1 and padj ≤ 0.01) are highlighted in blue. (c) Volcano plot of biotinylated proteins associated with H3K9me3 and H3K4me3 histone modification, as identified by mass spectrometry. Two of the known associated proteins are highlighted in red. The x-axis represents the log2 fold change and the y-axis represents −log10 p-value comparing three H3K9me3 AMPL-MS replicates with three H3K4me3 AMPL-MS replicates (paired). The significantly enriched proteins (LFC >1 and padj ≤ 0.01) are highlighted in blue. (d) Volcano plot of biotinylated proteins associated with H3K4me3 and H4K16ac histone modification, as identified by mass spectrometry and plotted as above (b). (e) Correlation plot showing similarity and reproducibility of AMPL-MS performed with H4K16ac_Milipore and H4K16ac_Active Motif; Pearson’s correlation (r) is reported in each block. (f) Volcano plot of biotinylated proteins associated with H4K16ac_Milipore and H4K16ac_Active Motif histone modification, as identified by mass spectrometry and plotted as (b).
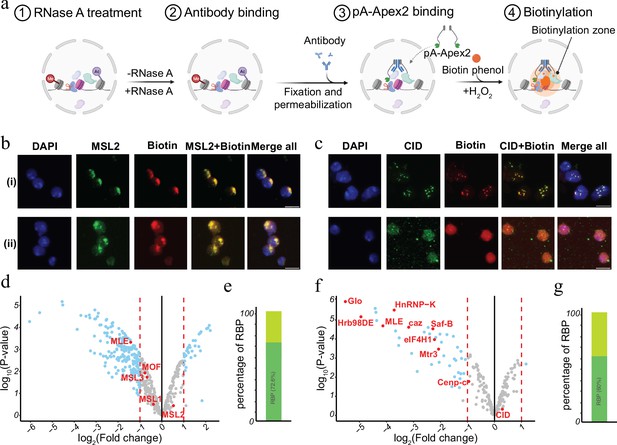
RNase treatment changes the proteomic environment of nuclear domains.
(a) Schematic of AMPL-MS-RNase method. Isolated nuclei were treated with RNase A, fixed, permeabilized, and incubated with specific antibodies. Recombinant pA-Apex2 enzyme binds to the antibody and biotinylates associated proximal proteins upon the addition of H2O2 and biotin-phenol. Created with BioRender.com. (b) Immunofluorescence of the X-chromosome-bound by an MSL2 antibody (in green) and biotinylated proteins after biotinylation by pA-Apex2 in control (i) and RNase A-treated samples (ii). Nuclei was stained by DAPI (in blue). Scale bars represent 5 µm. (c) Immunofluorescence of the centromer bound by an anti-CID antibody (in green) and biotinylated proteins after biotinylation by pA-Apex2 in control (i) and RNase A-treated samples (ii). Nuclei were stained by DAPI (in blue). Scale bars represent 5 µm. Volcano plot of proteins identified by AMPL-MS-RNase using an anti-MSL2 (d) or an anti-CID antibody (f). The bait protein is highlighted in red, along with components of the Drosophila dosage compensation complex. The x-axis represents the log2 fold change and the y-axis represents −log10 p-value comparing three control MSL-2 AMPL-MS replicates with three MSL2-RNase A-treated antibody-mediated proximity labelling coupled to mass spectrometry (AMPL-MS) replicates (paired). The significantly enriched proteins (LFC >1 and padj ≤ 0.01) are highlighted in blue. Percentage of RNA-sensitive proteins in proximity to MSL2 (e) or CID (f) containing known RNA-binding domains (RBP).
© 2024, BioRender Inc. Figure 4a was created using BioRender, and is published under a CC BY-NC-ND license. Further reproductions must adhere to the terms of this license.
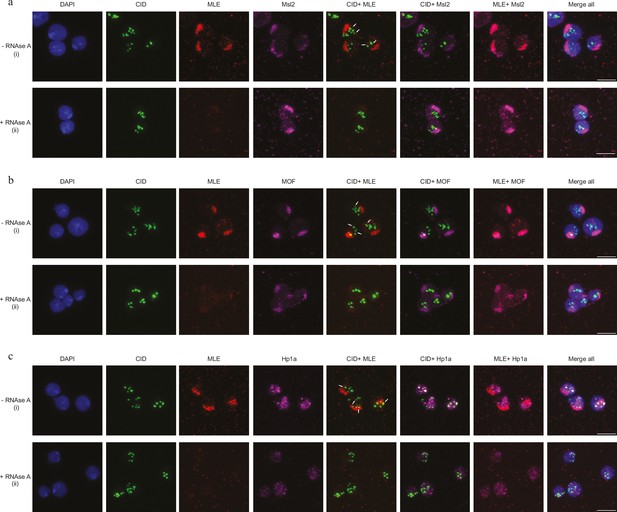
Maleless (MLE) associated with centromere in an RNA-dependent manner (related to Figure 4).
(a) Immunofluorescence image in control (i) and RNase A-treated samples (ii). Nuclei were stained by DAPI (in blue), CID (in green), MLE (in red), and Msl-2 (in magenta). Scale bars represent 5 μm. (b) Immunofluorescence image in control (i) and RNase A-treated samples (ii). Nuclei were stained by DAPI (in blue), CID (in green), MLE (in red), and MOF (in magenta). Scale bars represent 5 μm. (c) Immunofluorescence image in control (i) and RNase A-treated samples (ii). Nuclei were stained by DAPI (in blue), CID (in green), MLE (in red), and Hp1a (in magenta). Scale bars represent 5 μm.
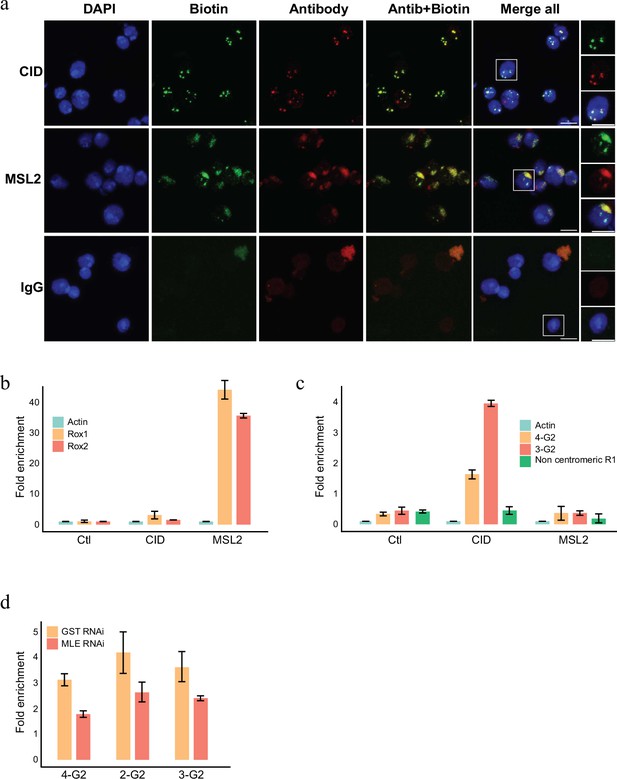
RNA labelling using antibody-mediated proximity labelling (AMPL) to study the RNA composition of chromatin domains.
(a) Immunofluorescence microscopy of centromeres and hyperactive X chromosome using CID and MSL2 antibody, respectively (in green), and the corresponding proximity RNA labelling after biotinylation by pA-Apex2 (in red). Nuclear DNA was stained by DAPI (in blue). IgG was used as antibody control. Scale bars represent 5 µm. (b) RT-qPCR analysis of enriched RNA in proximity to X chromosome. The PCR analysis is showing a specific enrichment of two long non-coding RNA Rox1 and Rox2 which are known to associate with X chromosome and actin as control. Relative abundance is calculated following 2−∆∆Ct method. Data are the mean of three replicates ±1 standard deviation (SD). (c) RT-qPCR analysis of enriched RNA in proximity to centromere. The PCR analysis is showing specific enrichment of RNA originates from centromeres 3 and 4 of SL2 cells. R1 is a control for non-centromeric transcript. Data are the mean of three replicates ±1 SD. (d) Enriched R-loop-ChIP-qPCR using hybrid binding domain (HBD) probe. The graph shows enrichment of R-loop in centromere for chromosomes 2–4 in control cells (gst RNAi) in comparison to MLE knock down cells. The enrichment is calculated relative to the input and is normalized by the Sdr promoter region as a non-centromeric control. Data are the mean of three replicates ±1 SD.
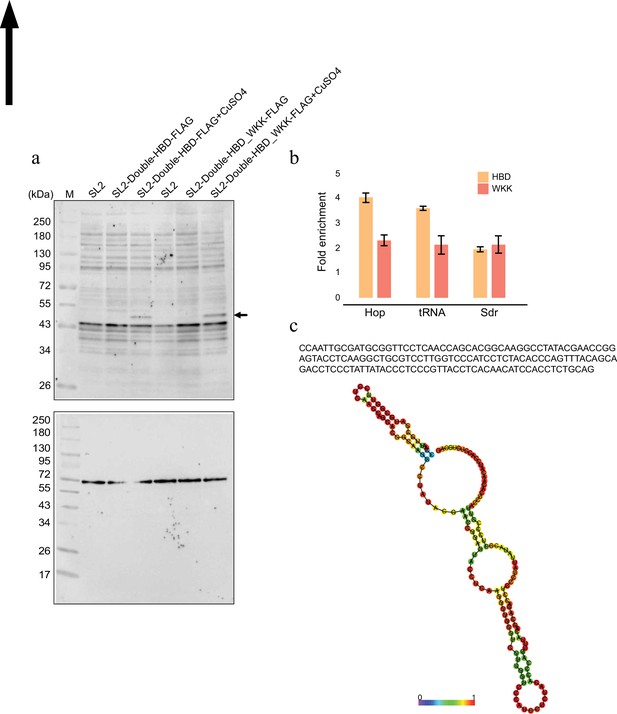
R-loop associated with centromere (related to Figure 5).
(a) Levels of expression of hybrid binding domain (HBD) and mutant (WKK) protein upon induction by CuSo4 analysed by western blotting using anti-FLAG antibody. Tubulin served as the loading control. (b) Enriched R-loop-ChIP-qPCR signals relative to input for validated R-loop-positive region (Hop and tRNA Lys) and negative region (Sdr) (Alecki and Francis, 2021) for HBD and WKK mutant. Results were calculated as the percentage of input and presented as mean ± standard error of the mean (SEM) (n = 3). (c) Sequence of the enriched transcript associated with centromere of chromosome 4. Secondary structure of the RNA as predicted by RNAfold (Gruber et al., 2008).
-
Figure 5—figure supplement 1—source data 1
Raw image files of the Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/95718/elife-95718-fig5-figsupp1-data1-v2.zip
-
Figure 5—figure supplement 1—source data 2
Labelled raw image files of the Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/95718/elife-95718-fig5-figsupp1-data2-v2.zip
Additional files
-
Supplementary file 1
CID-vs-AB: comparison of AMPL-MS proximity proteomics between CID antibody vs non-specific antibody as plotted in volcano plot Figure 1d.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp1-v2.xlsx
-
Supplementary file 2
ORA_CID-vs-AB: over representation analysis of protein identified in proximity to the centromeric chromatin domain (CID) as plotted in Figure 1f.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp2-v2.xlsx
-
Supplementary file 3
MSL2-vs-CID: comparison of AMPL-MS proximity proteomics between CID antibody vs Msl2 antibody as plotted in volcano plot Figure 2c.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp3-v2.xlsx
-
Supplementary file 4
ORA_MSL2-vs-CID: over representation analysis of protein identified in proximity to the centromeric chromatin domain (CID) vs hyperactive X chromosome (MSL2) as plotted in Figure 2e, f.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp4-v2.xlsx
-
Supplementary file 5
MSL2-vs-AB: comparison of AMPL-MS proximity proteomics between Msl2 antibody vs non-specific antibody.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp5-v2.xlsx
-
Supplementary file 6
AMPL-MS histone MS: analysis of histone modification associated with centromeric (CID) or hyperactive X chromosome (Msl2) as in Figure 2g.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp6-v2.xlsx
-
Supplementary file 7
Histone PTMs: comparison of proteomics data obtained for proteins associated with histone post-translational modifications (H3K9me3, H3K4me3, and H4K16Ac) as in Figure 3a,b,d and Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp7-v2.xlsx
-
Supplementary file 8
ORA_HistonePTMs: over representation analysis of protein found to be enriched in proximity to the H3K9me3 and H4K16Ac modifications as plotted in Figure 3c.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp8-v2.xlsx
-
Supplementary file 9
MSL2-RNase-vs-MSL2: a comparison of protein associated with hyperactive X chromosome with and without RNAse-A treatment as plotted in Figure 4d.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp9-v2.xlsx
-
Supplementary file 10
CID-RNase-vs-CID: a comparison of protein associated with centromere with and without RNAse-A treatment as plotted in Figure 4f.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp10-v2.xlsx
-
Supplementary file 11
CID_MSl2 RNase protein list: a list of protein found to be associated with centromere and hyperactive X chromosome.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp11-v2.xlsx
-
Supplementary file 12
Primer list: a list of primers used in the study.
- https://cdn.elifesciences.org/articles/95718/elife-95718-supp12-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95718/elife-95718-mdarchecklist1-v2.docx





