GDF2 and BMP10 coordinate liver cellular crosstalk to maintain liver health
Figures
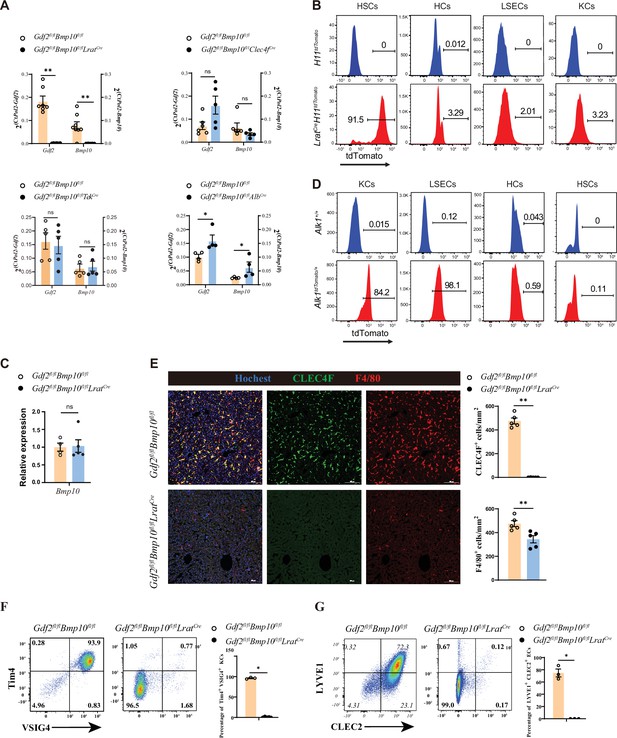
Hepatic stellate cells (HSCs) are the major source of GDF2 and BMP10 in the liver.
(A) Quantitative PCR analysis of Gdf2 and Bmp10 expression in the liver from the indicated mice at the age of 8–14 weeks (n=4–6/group). (B) Representative flow cytometric expression of tdTomato in HSCs, hepatocytes (HCs), endothelial cells (ECs), and Kupffer cells (KCs) from LratCreH11tdTomato mice at the age of 8–9 weeks (n=3/group). (C) Quantitative PCR analysis of Bmp10 expression in the right atrium from Gdf2fl/flBmp10fl/flLratCre and their littermate controls at the age of 8–12 weeks (n=4/group). (D) Representative flow cytometric expression of tdTomato in HSCs, HCs, ECs, and KCs from Alk1tdTomato mice at the age of 8–12 weeks (n=3/group). (E) Immunofluorescence images of liver sections from Gdf2fl/flBmp10fl/flLratCre and their littermate controls at the age of 3–12 weeks (n=6/group). Scale bars: 100 μm. (F) Flow cytometric expression of Tim4 and VSIG4 in KCs (pregated on CD45+Ly6C-F4/80+CD64+) and percentage of Tim4+VSIG4+ KCs from Gdf2fl/flBmp10fl/flLratCre (n=5) and their littermate controls (n=3) at the age of 8–12 weeks. (G) Flow cytometric expression of LYVE1 and CLEC2 in ECs (pregated on CD45+CD31-) and percentage of LYVE1+CLEC2+ ECs from Gdf2fl/flBmp10fl/flLratCre (n=3) and their littermate controls (n=3) at the age of 8–12 weeks. Results represent the mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001, by Mann-Whitney test (A, C, E–G).
-
Figure 1—source data 1
Numerical data of Figure 1A, C and E-G.
- https://cdn.elifesciences.org/articles/95811/elife-95811-fig1-data1-v1.xlsx
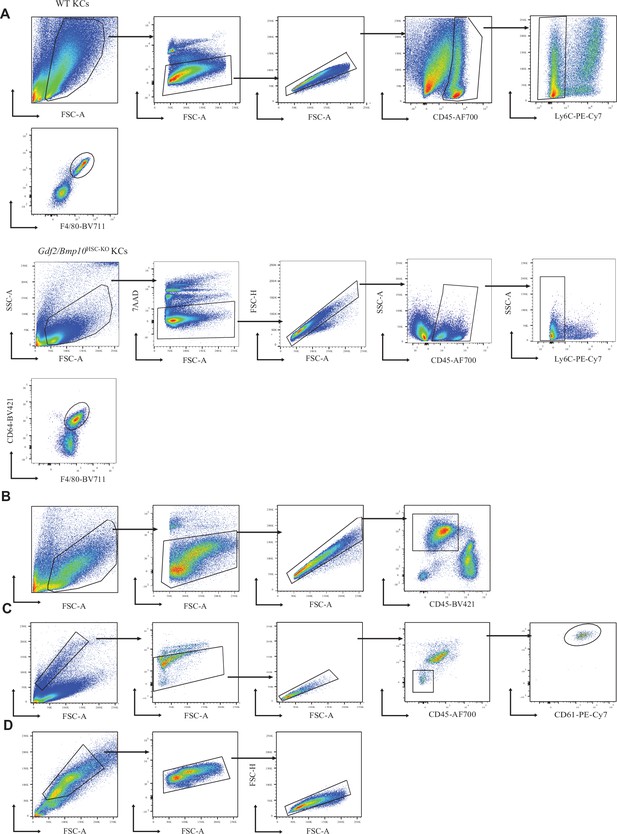
Gating strategies.
(A–D) Flow cytometry plots show representative pre-gating strategies for live CD45+Ly6C-CD64+F4/80+ Kupffer cells (KCs) from WT and Gdf2/Bmp10HSC-KO mice (A), live CD45-CD31+ endothelial cells (ECs) (B), live CD45-CD31-CD61+ hepatic stellate cells (HSCs) (C), and live SSChiFSChi hepatocytes (HCs) (D) in the liver.
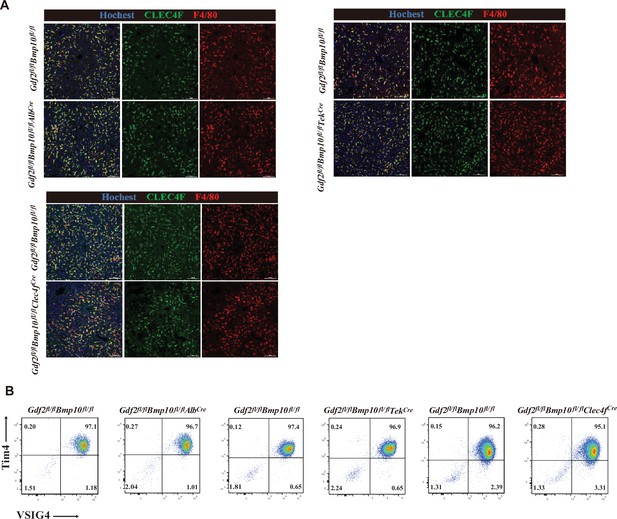
The phenotypes of Kupffer cells (KCs) were not affected in the liver from Gdf2fl/flBmp10fl/flAlbCre, Gdf2fl/flBmp10fl/flTekCre, and Gdf2fl/flBmp10fl/flClec4fCre mice.
(A) Representative immunofluorescence images of liver sections from the indicated mice at the age of 8–14 weeks (n=3–5/group). Scale bars: 100 μm. (B) Representative flow cytometric expression of Tim4 and VSIG4 in KCs from the indicated mice at the age of 8–12 weeks (n=3–5/group).
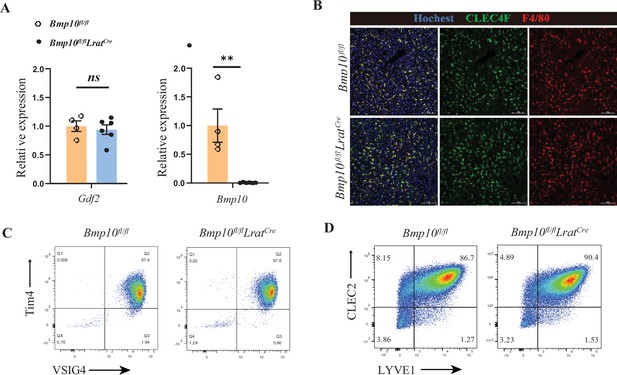
GDF2 can compensate the role of BMP10 in the liver.
(A) Quantitative PCR analysis of Gdf2 and Bmp10 expression in the liver from Bmp10fl/flLratCre and their littermate controls at the age of 12 weeks (n=4–6/group). (B) Representative immunofluorescence images of liver sections from the indicated mice at the age of 14 weeks (n=2/group). Scale bars: 100 μm. (C, D) Representative flow cytometric expression of the indicated markers in Kupffer cells (KCs) (C) and endothelial cells (ECs) (D) from Bmp10fl/flLratCre and their littermate controls at the age of 6–13 weeks (n=2–4/group). Results represent the mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001, by Mann-Whitney test (A).
-
Figure 1—figure supplement 3—source data 1
Numerical data of Figure 1—figure supplement 3A.
- https://cdn.elifesciences.org/articles/95811/elife-95811-fig1-figsupp3-data1-v1.xlsx
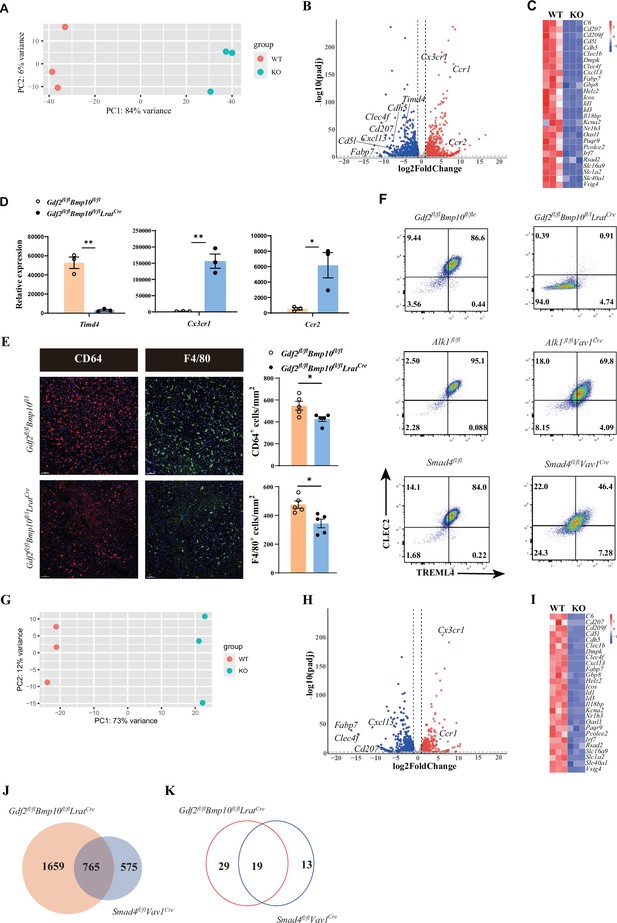
The differentiation of liver macrophages was inhibited in Gdf2fl/flBmp10fl/flLratCre mice.
(A, B) Principal component analysis (PCA) and volcano plot of the RNA-sequencing (RNA-seq) data of sorted CD45+Ly6C-F4/80+CD64+ liver macrophages from Gdf2fl/flBmp10fl/flLratCre and control mice at the age of 8–10 weeks (n=3/group). Genes upregulated and downregulated are shown in red and blue, respectively (fold change [FC]>2, adjusted p [p-adj]<0.05). (C) Heatmap showing signature genes expressed differentially in liver macrophages from Gdf2fl/flBmp10fl/flLratCre and control mice. (D) Expression counts of indicated genes in liver macrophages from Gdf2fl/flBmp10fl/flLratCre and control mice. ***p-adj<0.001. (E) Immunofluorescence images of F4/80+ and CD64+ liver macrophages in sections from Gdf2fl/flBmp10fl/flLratCre mice and their controls at the age of 12 weeks (n=5/group). Liver macrophages number was measured (right). Scale bars: 50μm. (F) Representative flow cytometric expression of CLEC2 and TREML4 in liver macrophages from the indicated mice at the age of 8–12 weeks (n=3–4/group). (G, H) PCA and volcano plot of the RNA-seq data of sorted liver macrophages from Smad4fl/flVav1Cre and control mice at the age of 8–10 weeks (n=3/group). Genes upregulated and downregulated are shown in red and blue, respectively (fold change [FC]>2, adjusted p [p-adj]<0.05). (I) Heatmap showing signature genes expressed differentially in liver macrophages from Smad4fl/flVav1Cre and control mice. (J, K) Venn diagram showing differentially expressed (DE) genes (J) and transcription factors (K) specific to liver macrophages of Gdf2fl/flBmp10fl/flLratCre, Smad4-deficient liver macrophages or shared between both mac populations. Results represent the mean ± SEM. *p<0.05, by Mann-Whitney test (E).
-
Figure 2—source data 1
Numerical data of Figure 2D and E.
- https://cdn.elifesciences.org/articles/95811/elife-95811-fig2-data1-v1.xlsx
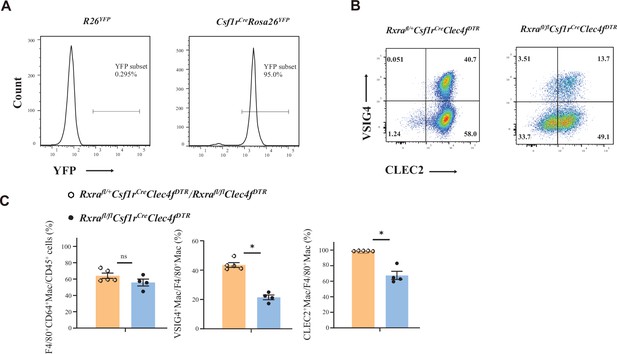
RXRα is required for the differentiation from blood monocytes to monocyte-derived Kupffer cells (MoKCs).
(A) Representative flow cytometric expression of YFP in blood monocytes from Csf1rCreRosa26YFP and control mice at the age of 8–12 weeks (n=2/group). (B, C) Representative flow cytometric expression (B) of CLEC2 and VSIG4 in liver macrophages, and percentage (C) of F4/80+, VSIG4+, and CLEC2+ liver macrophages from Rxrafl/flCsf1rCreClec4fDTR mice and controls at the age of 11–12 weeks. The indicated mice were injected with 200 ng/mice diphtheria toxin via IP. After 7 days, VSIG4 and CLEC2 expression in CD45+Ly6C-CD64+F4/80+ liver macrophages were assessed. Results represent the mean ± SEM. *p<0.05, by Mann-Whitney test (C).
-
Figure 2—figure supplement 1—source data 1
Numerical data of Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/95811/elife-95811-fig2-figsupp1-data1-v1.xlsx
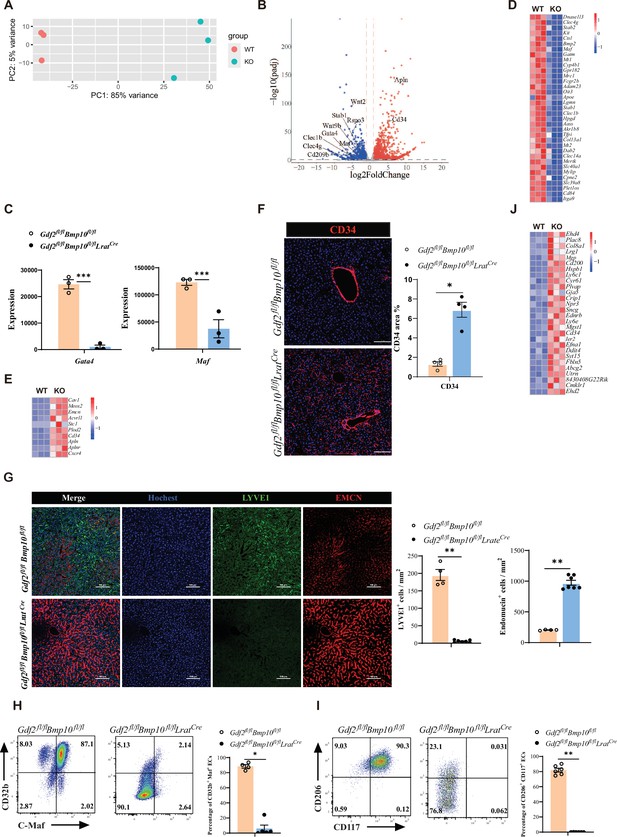
Endothelial cells (ECs) from Gdf2fl/flBmp10fl/flLratCre mice are transdifferentiated to continuous ECs.
(A, B) Principal component analysis (PCA) and volcano plot of the RNA-sequencing (RNA-seq) data of sorted hepatic ECs from Gdf2fl/flBmp10fl/flLratCre and control mice at the age of 8–10 weeks (n=3/group). Genes upregulated and downregulated are shown in red and blue, respectively (fold change [FC]>2, adjusted p [p-adj]<0.05). (C) Expression counts of Gata4 and Maf genes in ECs from Gdf2fl/flBmp10fl/flLratCre and control mice. ***p-adj<0.001. (D, E) Heatmap showing sinusoidal EC-associated genes (D) and continuous EC-associated genes (E) expressed differentially in hepatic ECs from Gdf2fl/flBmp10fl/flLratCre and control mice. (F) Representative immunofluorescence images in liver sections from Gdf2fl/flBmp10fl/flLratCre and control mice at the age of 12 weeks (n=4/group). CD34+ area was measured (right). Scale bars: 100 μm. (G) Representative immunofluorescence images in liver sections from Gdf2fl/flBmp10fl/flLratCre and control mice at the age of 8 weeks (n=4–7/group). LYVE1+ and EMCN+ area was measured (right). Scale bars: 100 μm. (H, I) Flow cytometric expression of CD32b, C-Maf, CD206, and CD117 in hepatic ECs from Gdf2fl/flBmp10fl/flLratCre and littermate controls at the age of 8–10 weeks (n=4–6/group). (J) Heatmap showing the indicated genes expressed differentially in hepatic ECs from Gdf2fl/flBmp10fl/flLratCre and control mice. Results represent the mean ± SEM. *p<0.05 and **p<0.01, by Mann-Whitney test (F–I).
-
Figure 3—source data 1
Numerical data of Figure 3C and F-I.
- https://cdn.elifesciences.org/articles/95811/elife-95811-fig3-data1-v1.xlsx
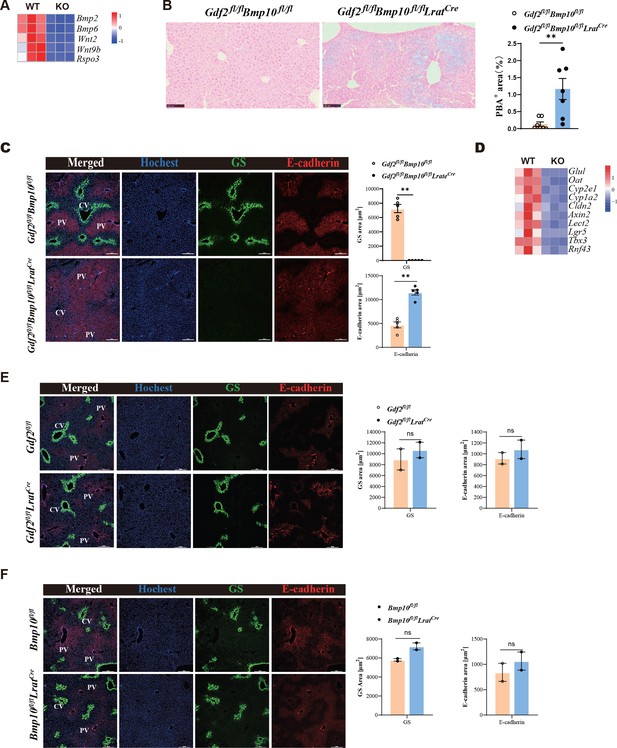
Liver metabolic zonation and iron metabolism are disrupted in Gdf2fl/flBmp10fl/flLratCre mice.
(A) Heatmap showing the indicated genes expressed differentially in hepatic endothelial cells (ECs) from Gdf2fl/flBmp10fl/flLratCre and control mice. (B) Prussian blue in livers from Gdf2fl/flBmp10fl/flLratCre mice and their controls at the age of 12 weeks (n=7/group). (C) Immunofluorescence images in liver sections from Gdf2fl/flBmp10fl/flLratCre mice and their controls at the age of 12 weeks (n=5/group). Scale bars: 200 μm. (D) Heatmap showing the indicated genes expressed differentially in liver tissues from Gdf2fl/flBmp10fl/flLratCre and control mice at the age of 12 weeks. (E, F) Immunofluorescence images in liver sections from Gdf2fl/flLratCre (E) and Bmp10fl/flLratCre (F) mice and their controls (n=2/group) at the age of 13–21 weeks (n=2/group). Scale bars: 200 μm. Results represent the mean ± SEM. **p<0.01, by Mann-Whitney test (B, C, E, F).
-
Figure 4—source data 1
Numerical data of Figure 4B, C, E and F.
- https://cdn.elifesciences.org/articles/95811/elife-95811-fig4-data1-v1.xlsx
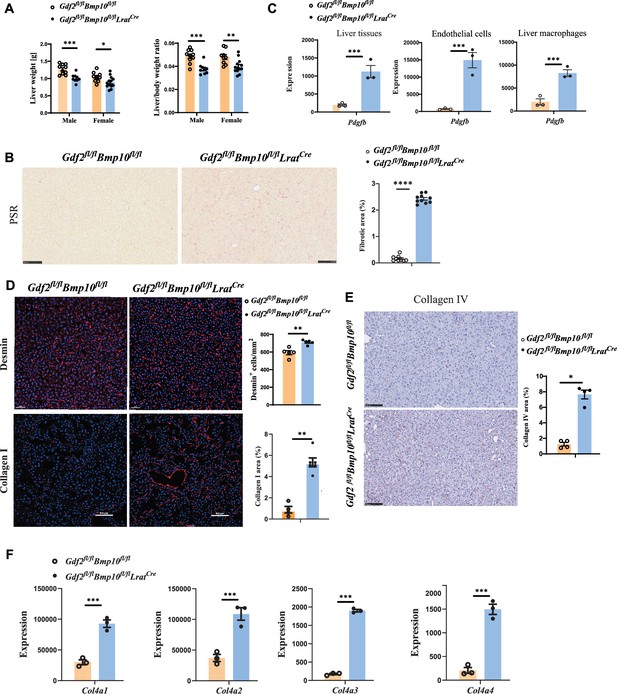
Liver fibrosis occurs in Gdf2fl/flBmp10fl/flLratCre mice.
(A) Liver weight and liver to body weight ratio of Gdf2fl/flBmp10fl/flLratCre mice and their littermate controls at the age of 12 weeks. (B) Picrosirius red (PSR) staining of liver sections from Gdf2fl/flBmp10fl/flLratCre mice and their controls at the age of 12 weeks (n=9–10/group). Scale bars: 100 μm. (C) Expression counts of Pdgfb gene in liver tissues (left), endothelial cells (middle), and liver macrophages (right) from Gdf2fl/flBmp10fl/flLratCre mice and their control mice. ***p-adj<0.001. (D) Immunofluorescence images of Desmin and collagen I in liver sections in liver tissues from Gdf2fl/flBmp10fl/flLratCre and control mice at the age of 12–28 weeks (n=4–6/group). Scale bars: 50μm (upper) and 100μm (lower). (E) Collagen IV immunohistochemistry staining of liver sections from Gdf2fl/flBmp10fl/flLratCre mice and their controls at the age of 12 weeks (n=4–5/group). Scale bars: 100 μm. (F) Expression counts of indicated genes in endothelial cells from Gdf2fl/flBmp10fl/flLratCre and control mice. ***p-adj<0.001. Results represent the mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001, by Mann-Whitney test (A, B, D, E).
-
Figure 5—source data 1
Numerical data of Figure 5A-F.
- https://cdn.elifesciences.org/articles/95811/elife-95811-fig5-data1-v1.xlsx
Additional files
-
Supplementary file 1
Transcription factors (TFs) downregulated in liver macrophages from Gdf2/Bmp10HSC-KO mice and Smad4fl/fl Vav1Cre mice.
- https://cdn.elifesciences.org/articles/95811/elife-95811-supp1-v1.xlsx
-
Supplementary file 2
Primers used for real-time PCR.
- https://cdn.elifesciences.org/articles/95811/elife-95811-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95811/elife-95811-mdarchecklist1-v1.docx





