A ‘torn bag mechanism’ of small extracellular vesicle release via limiting membrane rupture of en bloc released amphisomes (amphiectosomes)
Figures
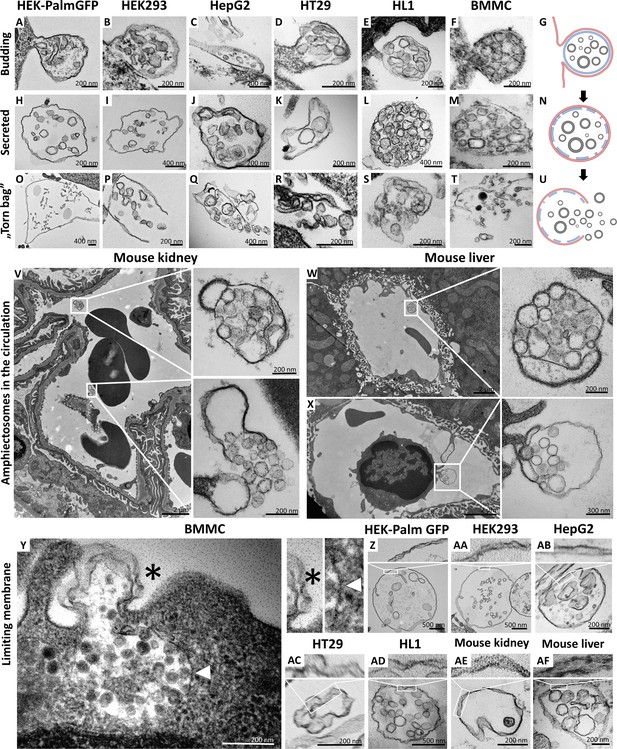
Transmission electron microscopic detection of the release and extracellular fate of large, multivesicular extracellular vesicles (MV-lEVs) secreted by different cell lines and cells in mouse organs.
Major steps of the release of MV-lEVs were detected in the case of all tested cell lines including the immortal, non-tumorous HEK293T-PalmGFP (A, H, O), HEK293 (B, I, P), the tumorous cell lines HepG2 (C, J, Q) and HT29 (D, K, R), the beating cardiomyocyte cell line HL1 (E, L, S) and the primary suspension of bone marrow-derived mast cells (BMMCs) (F, M, T). The different phases of EV secretion were also captured in the circulation of mouse kidney (V) and liver (W, X). According to the electron micrographs, we found evidence for the budding (A–G, X) and secretion (H–N, V, W) of the MV-lEVs. We also detected the extracellular rupture of the limiting membrane of the released MV-lEVs with the escape of the intraluminal vesicles (ILVs) by a ‘torn bag mechanism’ (O–U, V). Although it is not always clear whether the secreted MV-lEVs have a single or double limiting membrane, several micrographs suggest the presence of the double membrane (Y–AF) in the secreted MV-lEVs. In the case of BMMCs (Y), the release phase of a multivesicular structure is captured. The bottom portion of this structure embedded in the cytoplasm is surrounded by a single membrane (white arrowhead) while the upper (budding) portion is covered by double membrane (asterisk). In the schematic figures (G, N, U) the limiting membrane of MV-lEV presumably with plasma membrane origin was indicated by red, the original limiting membrane of intracellular amphisomes, which may be fragmented during the release process was indicated by blue while the ILVs of the MV-lEV were shown by gray color. Panel G was created with BioRender.com. Panel N was created with BioRender.com. Panel U was created with BioRender.com.
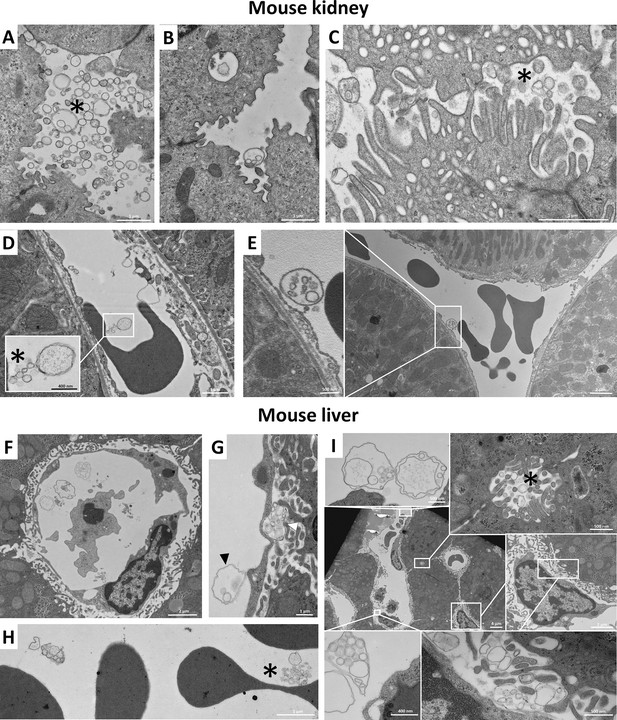
Additional transmission electron micrographs of mouse kidney and liver sections.
For further evaluation of the presence of different extracellular vesicles (EVs) in mouse kidney (A–E) and liver (F–I), low and high magnification images are shown. Presence of individual small and large EVs is indicated by asterisk in the case of mouse kidney (A, C, D) and mouse liver (F, H, I). The multivesicular large EVs (MV-lEVs) and individual EVs were present simultaneously (C, D, F, H, I). In the mouse liver ultrathin section (G), MV-lEV secretion by endothelial and subendothelial cells (black and white arrow heads, respectively) were detectable in the same image.
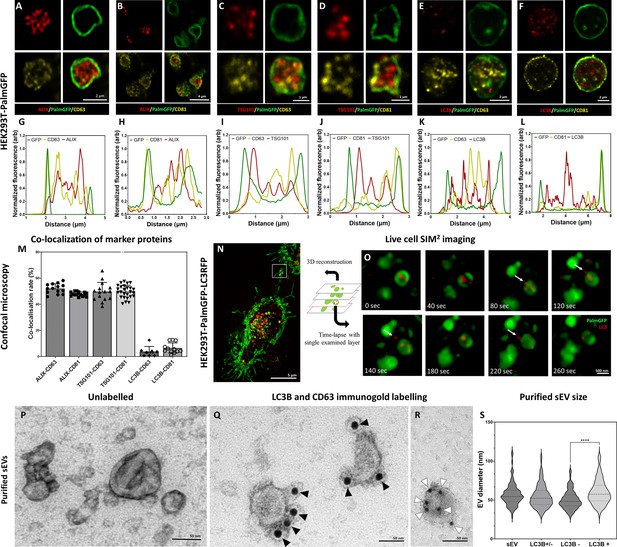
Detection of conventional small extracellular vesicle (sEV) markers and the LC3 protein in HEK293T-PalmGFP cell-derived EVs.
Widely used sEV markers (CD63, CD81, ALIX, and TSG101) and LC3B were tested in multivesicular large EVs (MV-lEVs) found in the microenvironment of the releasing cells by confocal microscopy after in situ fixation (A–F). Normalized fluorescence intensities were calculated to determine the relative localization of the limiting membrane (PalmGFP), the conventional sEV markers, and the LC3B signal (G–L). Fluorescence intensity peaks of sEV markers were largely overlapping with each other, while the LC3B signal and the sEV markers showed separation. Co-localization rates were also calculated (M). The sEV markers co-localized with one another as no significant difference was found among them. In contrast, low co-localization rates were detected between the ‘classical’ sEV markers and LC3B (one-way ANOVA, p<0.0001, n=8-26 confocal images). Error bars represent standard deviation. Real-time release of LC3 positive sEVs by the ‘torn bag mechanism’ was studied in the case of HEK293T-PalmGFP-LC3RFP cells by Elyra7 SIM2 super-resolution live-cell imaging (N,O). Images were recorded continuously and selected serial time points are shown. LC3 positive, red fluorescent small particles were released within a 5 min timeframe (O) and are indicated by white arrows. Presence of CD63 and LC3B were detected in the case of an sEV fraction separated from serum-free condition medium using immunogold transmission electron microscopy (TEM). HEK293T-PalmGFP-derived sEV fraction is shown by negative-positive contrast without immune labeling (P). In double-labeled immunogold TEM images (Q, R), distinct LC3B positive (Q) and CD63 positive (R) sEVs were found. However, CD63-LC3B double positive EVs were not detected. Black arrowheads indicate 10 nm gold particles identifying LC3B, while white arrowheads show 5 nm gold particles corresponding to the presence of CD63. Quantitative analysis of TEM images was performed (S), and the diameters of different EV populations were determined. The LC3B negative population was significantly smaller than the LC3B positive one (p<0.0001, t-test; n=79–100). No difference was detected when the immunogold labeled sEV fraction (either LC3B positive or negative, LC3B+/-) and the unlabeled sEV fraction (sEV) were compared (p<0.05, t-test, n=112–179).
-
Figure 2—source data 1
XLSX file containing data points of Figure 2G, H, I, J, K, L, M, and S.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-data1-v1.xlsx

Localization of GFP signal in HEK293T-PalmGFP cells.
The GFP positivity of the HEK293TPalmGFP cells was controlled by immunofluorescence microscopy on control (A) and 30 µM Chloroquine-treated (B) cells. Co-localization of PalmGFP and the anti-GFP signal was calculated in the plasma membrane and in the cytoplasm (C) and Pearson’s correlations were visualized. The green fluorescence in the plasma membrane was clearly GFP-dependent in the case of control (Ctrl Membrane) and Chloroquine-treated (Chloro Membrane) samples, while in the cytoplasm, the correlation was significantly weaker (Ctrl Cytoplasm and Chloro Cytoplasm). Within the cytoplasm, the correlation was stronger in Chloroquine-treated cells, suggesting that endosomal membrane may contain PalmGFP possibly as a result of the membrane endocytotic or recycling processes (p<0.0001, t-test; n = 45). Error bars represent standard deviation.
-
Figure 2—figure supplement 1—source data 1
XLSX file containing data points of Figure 2-figure supplement 1C.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-figsupp1-data1-v1.xlsx
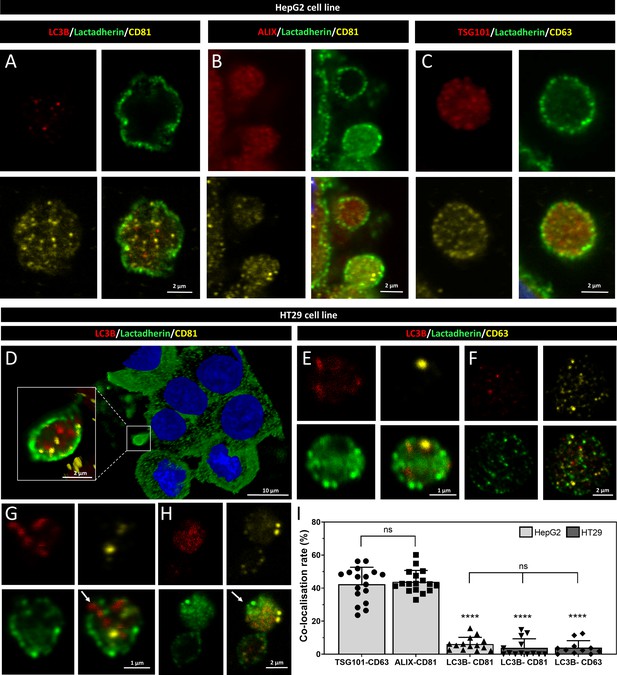
Confocal microscopic images of amphiectosome release by HT29 and HepG2 cells.
The intraluminal extracellular vesicles (EVs) of multivesicular large EVs (MV-lEVs) were found to be positive for LC3B and CD81 in HepG2 (A) and HT29 cells (D). The presence of ALIX/CD81 (B) and TSG101/CD63 (C) was also examined in the released amphiectosomes of the HepG2 cell line. In the case of HT29 cells, phases of the ‘torn bag mechanism’ were captured, including a secreted intact amphiectosome (E), amphiectosomes with ruptured limiting membrane releasing internal vesicles (G, white arrow), an inside-out secreted amphiectosome (H, white arrow), and an amphiectosome with a fully disintegrated limiting membrane and released small EVs (sEVs) (F). Co-localization rates of marker proteins were calculated (I). The typical sEV markers of HepG2 co-localized with each other. A significant difference was not found among them. In contrast, low co-localization rates were detected between the ‘classical’ sEV markers (CD63 and CD81) and LC3B. The co-localization rates between ‘classical’ sEV markers (LC3B) and within ‘classical’ EV markers (TSG101-CD63 and ALIX-CD81) were found significantly different (one-way ANOVA, p<0.0001, n=11–17 confocal images). Error bars represent standard deviation.
-
Figure 2—figure supplement 2—source data 1
XLSX file containing data points of Figure 2—figure supplement 2I.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-figsupp2-data1-v1.xlsx
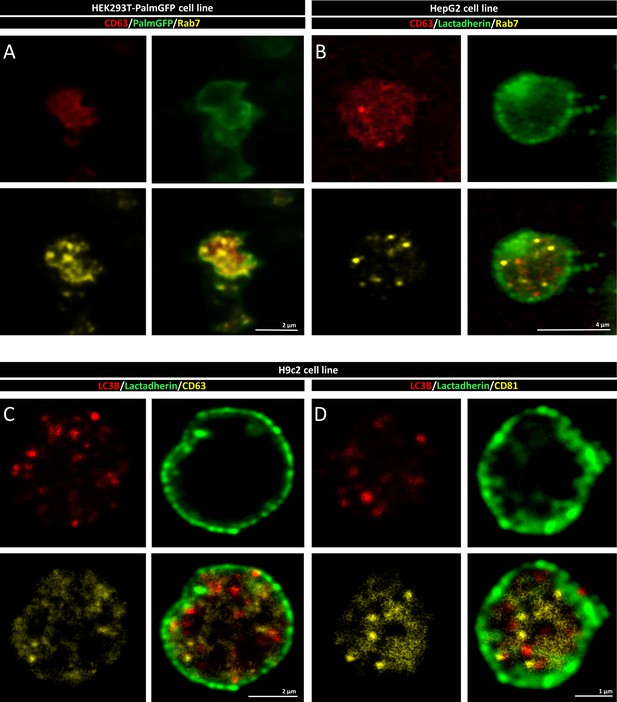
Additional confocal microscopic images of H9c2, HEK293T-PalmGFP, and HepG2 cells-derived multivesicular large extracellular vesicles (MV-lEVs).
Released amphiectosomes of H9c2 rat cardiomyoblast cell line were captured. They contain CD63, CD81, or LC3B positive intraluminal vesicles (ILVs) (A, B). The MV-lEVs released by HEK293T-PalmGFP (C) and HepG2 cells (D) were tested for CD63 and the Rab7 late endosomal marker. Both CD63 and Rab7 were present in association with the ILVs.
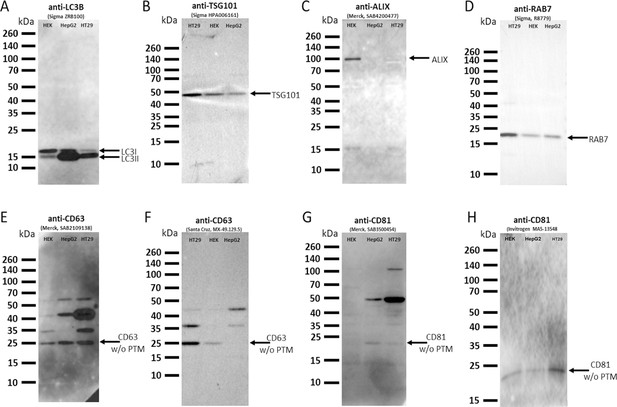
Qualitative western blot validation of antibodies used in immunofluorescence detection.
Whole-cell lysates from three distinct human cell lines (HEK293, HepG2, and HT29) were employed in the validation process to reduce the cell line-specific variations in this qualitative study. Protein bands lacking posttranslational modifications are noted as ‘w/o PTM’. The found posttranslational modifications of CD63 and CD81 are widely and well recognized, the results are in agreement with the western blot data provided by the antibody suppliers.
-
Figure 2—figure supplement 4—source data 1
Original files for western blot analysis displayed in Figure 2—figure supplement 4.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-figsupp4-data1-v1.zip
-
Figure 2—figure supplement 4—source data 2
Original western blots for Figure 2—figure supplement 4, indicating the relevant bands and cell lines.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-figsupp4-data2-v1.zip
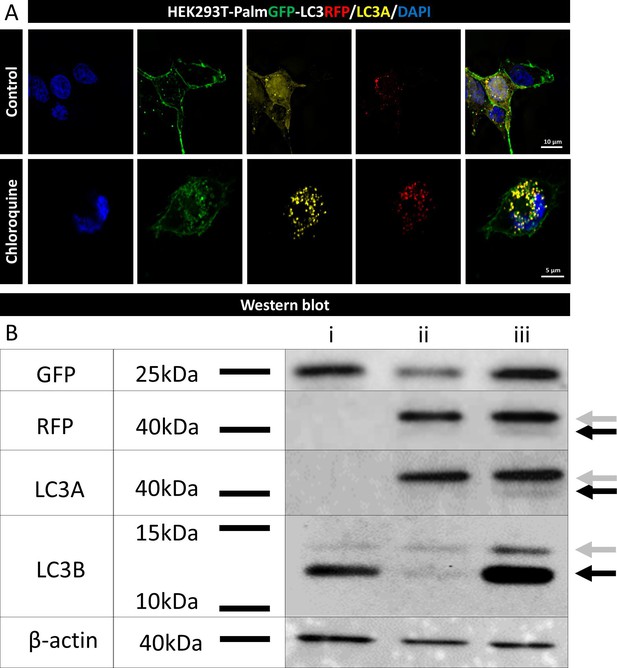
Characterization of the in-house generated HEK293T-PalmGFP-LC3RFP cell line.
Confocal microscopy (A) and western blot analysis (B) were employed to characterize the HEK293T-PalmGFP-LC3RFP cell line. For overnight Chloroquine treatment, 30 μM Chloroquine was applied. Punctate LC3 fluorescence observed in (A) corresponds to autophagosomes or amphisomes. In panel (B), whole-cell lysates from HEK293T-PalmGFP (lane i), HEK293T-PalmGFP-LC3RFP (lane ii), and Chloroquine-treated HEK293T-PalmGFP-LC3RFP (lane iii) samples were examined. Gray arrows indicate LC3I, while black arrows highlight the lipidated LC3II.
-
Figure 2—figure supplement 5—source data 1
Original files for western blot analysis displayed in Figure 2—figure supplement 5B.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-figsupp5-data1-v1.zip
-
Figure 2—figure supplement 5—source data 2
Original western blots for Figure 2—figure supplement 5B, indicating the relevant bands, cell lines, and treatments.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-figsupp5-data2-v1.zip
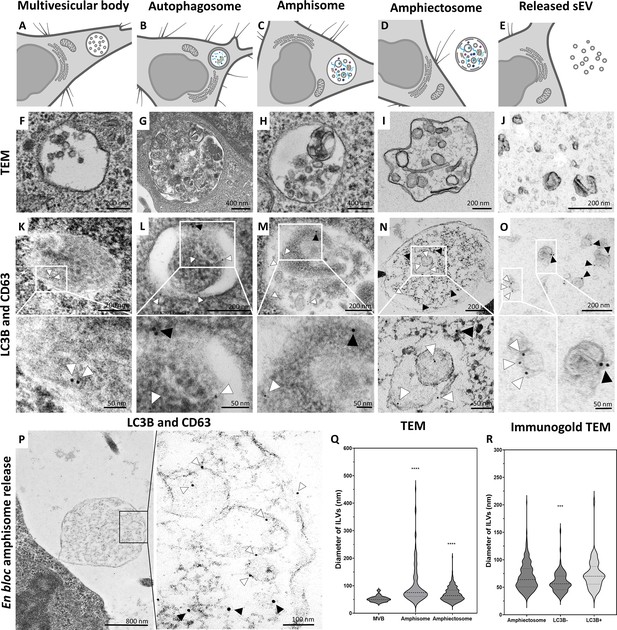
Structures involved in amphiectosome release.
Multivesicular body (MVB, A), autophagosome (B), amphisome (C), amphiectosome (D), and secreted small extracellular vesicles (sEVs) (E) were identified by transmission electron microscopy (TEM) with and without immunogold labeling of HEK293T-PalmGFP cell cultures. Panel A was created with BioRender.com. Panel B was created with BioRender.com. Panel C was created with BioRender.com. Panel D was created with BioRender.com. Panel E was created with BioRender.com. White arrowheads (5 nm gold particles) indicate CD63 and black arrowheads (10 nm gold particles) show LC3B. While MVBs (F) were LC3B negative (K), we detected CD63 positivity on the surface of the intraluminal vesicles (ILVs) (K). In an autophagosome (G), the limiting membrane layers were positive for CD63 and LC3B (L). In contrast, the internal membranes of autophagosome were CD63 single positive (L). In the case of an amphisome (H), heterogeneous membrane structures were visible with variable size and morphology. The ILVs were either CD63 or LC3B positive (M). The amphiectosomes were located in the extracellular space and contained ILVs of different size and shape (I). The ILVs of amphiectosome (as in case of amphisome) were either CD63 or LC3B positive (N). Secreted sEVs purified from serum-free conditioned medium (J) with immunogold labeling (O) were also found to be either CD63 or LC3B positive. Release of an amphiectosome is shown (P) with CD63 and LC3B immunogold signals. Higher magnification of the insert is indicated by the black rectangle. It shows either CD63 or LC3B positive ILVs. Size distributions of ILVs of MVBs, amphisomes, and amphiectosomes were determined on Epon-embedded ultrathin sections (Q). Although the ILV sizes differed significantly (one-way ANOVA, ****: p<0.0001, n=73, 138, and 595, respectively), the majority of ILVs had a diameter between 40 and 100 nm. The diameter of LC3B positive and negative ILVs of amphiectosomes was assessed on TEM images of immunogold labeled ultrathin sections (R). LC3B negative ILVs were significantly smaller than the LC3B positive ones, while the ILVs in the Epon-embedded sections did not differ from the LC3B positive ones (one-way ANOVA, p<0.001, n = 595, 101, and 70, respectively).
-
Figure 2—figure supplement 6—source data 1
XLSX file containing data points of Figure 2—figure supplement 6Q and R.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig2-figsupp6-data1-v1.xlsx
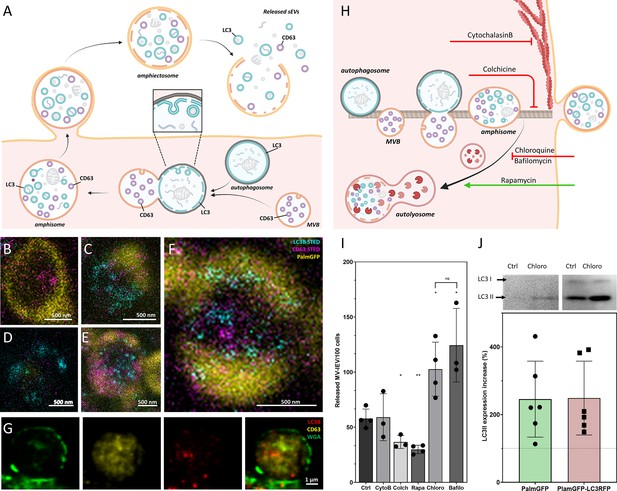
Amphiectosome release and its modulation.
Based on our data, a model of amphiectosome release was generated (A). Panel A was created with BioRender.com. According to this model, the fusion of multivesicular bodies (MVBs) and autophagosomes forms amphisomes. The LC3B positive membrane layer (indicated in cyan) undergoes disintegration and forms LC3B positive intraluminal vesicles (ILVs) inside the amphisome. Later, the amphisome is released into the extracellular space by ectocytosis and can be identified extracellularly as an amphiectosome. Finally, the limiting membrane(s) of the amphiectosome is ruptured and the ILVs are released as small extracellular vesicles (sEVs) into the extracellular space by a ‘torn bag mechanism’. Steps of amphisome formation including LC3 positive ILV formation in 30 µM Chloroquine-treated HEK293T-PalmGFP cells was followed by super-resolution (stimulated emission depletion [STED]) microscopy (B–F). The super-resolution STED channels were LC3B (cyan) and CD63 (magenta), while yellow indicates the confocal PalmGFP signal. Intracellular vesicular structures (such as endosomes, MVBs, and amphisomes) may receive PalmGFP from the plasma membrane. An MVB (B), an autophagosome with PalmGFP negative membrane (D), fusion of an autophagosome and an MVB (C), formation of LC3B positive ILVs in an amphisome (F), and a mature amphisome (E) were detected. To confirm the origin of the external membrane layer of amphiectosomes, fluorescently labeled wheat germ agglutinin (WGA) was applied. The plasma membrane of the living non-fluorescent HEK293 cells was labeled. As the external membrane of the budding amphiectosome was WGA positive, its plasma membrane origin is confirmed (G). To further support our model on amphiectosome release and ‘torn bag’ EV secretion, different in vitro treatments were applied. Cytochalasin B, Colchicine, Chloroquine, Bafilomycin A1, and Rapamycin were used to modulate amphiectosome release. Targeted molecular processes are summarized (H). Panel H was created with BioRender.com. While Cytochalasin B inhibits actin-dependent membrane budding and cell migration, Colchicine blocks the microtubule-dependent intracellular trafficking. While Chloroquine and Bafilomycin have similar, Rapamycin has opposite effect on lysosome-autophagosome or lysosome-amphisome fusion. Chloroquine and Bafilomycin inhibit lysosomal degradation while Rapamycin accelerates it. Based on confocal microscopy, Cytochalasin B (CytoB) did not alter the dynamics of amphiectosome release (I). In contrast, both Colchicine (Colch) and Rapamycin (Rapa) significantly inhibited the release of amphiectosomes, while Chloroquine (Chloro) and Bafilomycin (Bafilo) increased the release frequency. There was no difference between the effect of Chloroquine and Bafilomycin (I). Results are shown as mean ± SD of three to four independent biological replicates, analyzed by one-way ANOVA and Student’s t test, *: p<0.05, **: p<0.01, ns: non-significant. Original LASX files, which served as a basis of our quantification, are publicly available (doi: 10.6019/S-BIAD1456). Example for the calculation is shown in Figure 3—figure supplement 1H. Presence of membrane-bound (lipidated) LC3II was tested by western blotting. The total protein content of serum-, cell-, and large EV-depleted conditioned medium of HEK293T-PalmGFP (PalmGFP) and HEK293T-PalmGFP-LC3RFP (PalmGFP-LC3RFP) cells was precipitated by TCA and 20 µg of the protein samples were loaded on the gel (J). The lipidated LC3II band was detected in all cases. Relative expression of control (Ctrl) and Chloroquine (Chloro)-treated samples were determined by densitometry. Chloroquine treatment increased the LC3II level by approximately twofold. Results are shown as mean ± SD of n=6 biological replicates.
-
Figure 3—source data 1
XLSX file containing data points of Figure 2I and J.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Original files for western blot analysis displayed in Figure 3J.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig3-data2-v1.zip
-
Figure 3—source data 3
Original western blots for Figure 3J, indicating the relevant bands, cell lines, and treatments.
- https://cdn.elifesciences.org/articles/95828/elife-95828-fig3-data3-v1.zip
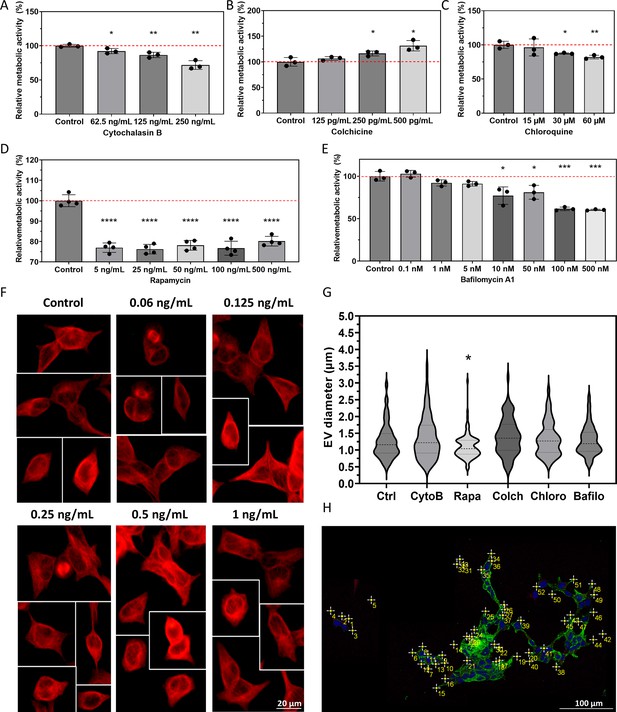
Supporting information for treatments and size distribution of multivesicular large extracellular vesicles (MV-lEVs).
Relative metabolic activity was assessed through a Resazurin assay (A–E) during treatment optimization. The red dashed line indicates 100% metabolic activity, representing control cells. Results are presented as mean ± SD values of n=3–4 independent biological replicates. Student’s unpaired t-test was performed to compare control and treated cells (*: p<0.05, **: p<0.01, ****: p<0.0001). For Colchicine treatment, alterations in the microtubular network were observed through immunocytochemistry (F) and documented using an epifluorescent microscope. Changes in the diameter of released MV-lEVs under various treatments were determined on confocal microscopy images. A significant reduction in size was identified only in the case of Rapamycin treatment (G, one-way ANOVA test *: p<0.05, n=95–101). An example for MV-lEV number calculation based on confocal images is shown (H).
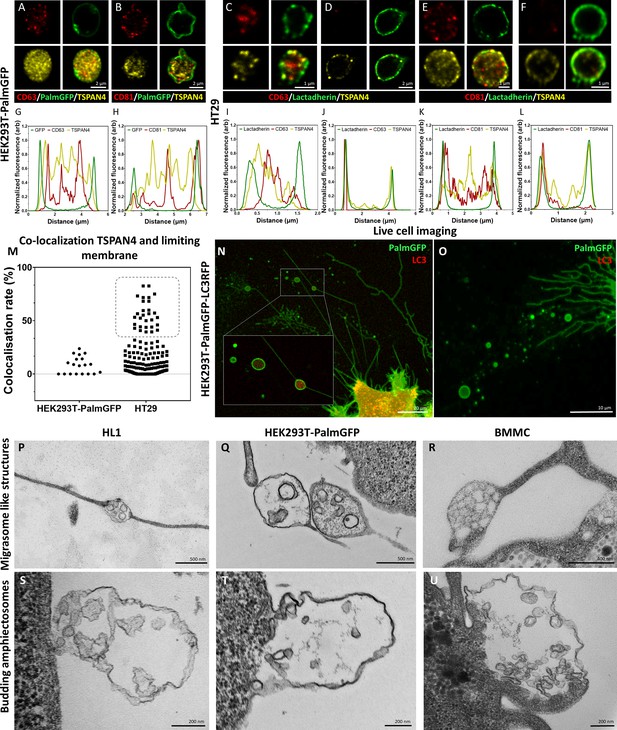
Comparison of amphiectosomes and migrasomes.
Commonly used small extracellular vesicle (sEV) markers (CD63, CD81) and TSPAN4, a suggested migrasome marker, were tested in in situ fixed intact multivesicular large EVs (MV-lEVs) of HEK293T-PalmGFP (A, B) and HT29 (C–F) cells by confocal microscopy. Normalized fluorescence intensities were calculated to determine the relative localization of the limiting membrane (with PalmGFP or lactadherin labeling) and the CD63/TSPAN4 and CD81/TSPAN4 markers (G–L). In the case of HEK293T-PalmGFP-derived EVs, we did not find migrasomes with TSPAN4 in their limiting membrane. The TSPAN4 signal was only detected intraluminally in the MV-lEVs. The limiting membranes of HT29-derived MV-lEVs were either TSPAN4 positive or negative. The co-localization rate between the limiting membrane and TSPAN4 was low in case of HEK293T-PalmGFP-derived EVs. In the case of HT29 cells, two MV-lEV populations were identified: one with low and one with high co-localization rates (M). Live-cell imaging of HEK293T-PalmGFP-LC3RFP cells showed retraction fiber-associated MV-lEVs with or without intraluminal LC3 positivity (N, O). Using transmission electron microscopy (TEM), we could identify structures with retraction fiber-associated morphology in the case of HL1 cells (P), HEK293T-PalmGFP cells (Q), and bone marrow-derived mast cells (BMMCs) (R). For comparison, budding of amphiectosomes of the same HL1 cells (S), HEK293TPalmGFP cells (T), and BMMCs (U) are shown (without being associated with long retractions fibers).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293 human kidney(embryonic) | ECACC (Sigma) | #85120602 RRID:CVCL_0045 | Batch No: 18E026 |
| Cell line (Homo sapiens) | HT29 Caucasian colon adenocarcinoma grade II | ECACC (Sigma) | #91072201 RRID:CVCL_0320 | Batch No: 09K003 |
| Cell line (Homo sapiens) | HepG2 human hepatocyte carcinoma | ECACC (Sigma) | #85011430 RRID:CVCL_0027 | Batch No: 19B009 |
| Cell line (Mus musculus) | HL1 mouse cardiomyocyte cell line, atrial | Merck | # SCC065 RRID:CVCL_0303 | Batch No: RD1601001 |
| Cell line (Homo sapiens) | HEK293TPalmGFP human kidney(embryonic) expressing palmitoylated GFP | Kind gift of Charles Lai https://doi.org:10.103 8/ncomms8029 | Resorted before MCB preparation | |
| Cell line (Homo sapiens) | HEK293T- PalmGFPLC3RFP human kidney(embryonic) expressing palmitoylated GFP and RFP tagged LC3 | This paper | See Materials and methods | |
| Cell line (Mus musculus) | BMMC bone marrow-derived mast cells | Primary cell culture https://doi.org/10.1 002/jev2.12023 | ||
| Cell line (Rattus norvegicus) | H9C2 (2-1) rat cardiovascular , Myoblast | ECACC (Sigma) | #88092904, RRID:CVCL_0286 | Batch No: 17A028 |
| Transfected construct | LentiBrite RFP-LC3 Lentiviral Biosensor | Merck | 17-10143 | Batch No: 3530171 |
| Biological sample (Mus musculus) | own animal house | C57BL/6 RRID:MGI:2159769 | male, 12 weeks of age | |
| Antibody | rabbit polyclonal anti-CD63 (Cterminal) | Sigma/Merck | SAB2109138 | IF (1:200) WB (1:500) |
| Antibody | mouse monoclonal anti-CD63 | Santa Cruz Biotechnology | MX-49.129.5 clone: sc-5275 RRID:AB_627877 | IF (1:200) TEM (1:50) WB (1:1000) |
| Antibody | rabbit polyclonal anti-CD81 | Sigma/Merck | SAB3500454 RRID:AB_10640751 | IF (1:200) WB (1:2500) |
| Antibody | mouse monoclonal anti-CD81 | Invitrogen | MA5-13548 clone: 1.3.3.22 RRID:AB_10987151 | IF (1:100) WB (1:100) |
| Antibody | rabbit polyclonal anti-TSG101 | Sigma/Merck | HPA006161 RRID:AB_1080408 | IF (1:200) WB (1:1000) |
| Antibody | rabbit polyclonal anti-ALIX (Cterminal) | Sigma/Merck | SAB420047 | IF (1:200) WB (1:1000) |
| Antibody | rabbit monoclonal anti-LC3B | Sigma/Merck | ZRB100 clone: 12K5 | IF (1:200) TEM (1:50) WB (1:1000) |
| Antibody | rabbit monoclonal anti-LC3A | Sigma/Merck | ZRB1125 clone: 3J12 | IF (1:200) WB (1:1000) |
| Antibody | rabbit polyclonal anti-TSPAN4 | Bioss | BS-9413R | IF (1:200) |
| Antibody | mouse monoclonal anti-Rab7 | Sigma/Merck | R8779 clone: Rab7117 RRID:AB_609910 | IF (1:200) WB (1:1000) |
| Antibody | mouse monoclonal anti-α-tubulin | Sigma/Merck | T9026 clone: DM1A RRID:AB_477593 | IF (1:200) |
| Antibody | mouse monoclonal anti-GFP | Sigma/Merck | G6539 clone: GFP-20 RRID:AB_259941 | IF (1:200) WB (1:1000) |
| Antibody | mouse monoclonal anti-RFP | Invitrogen | MA5-15257 clone: RF5R RRID:AB_10999796 | WB (1:1000) |
| Antibody | goat antimouse IgGATTO550 | Sigma/Merck | 43394 RRID:AB_1137651 | IF (1:1000) |
| Antibody | goat antirabbit IgG- ATTO647N | Sigma/Merck | 40839 RRID:AB_1137669 | IF (1:1000) |
| Antibody | goat antimouse Star 635P | Abberior | ST635P-1001–500UG RRID:AB_2893232 | IF (1:500) |
| Antibody | goat antirabbit Star 580 | Abberior | ST580-1002-500UG RRID:AB_2910107 | IF (1:500) |
| Antibody | goat polyclonal anti-rabbit IgG Fc (HRP) | abcam | ab97200 RRID:AB_10679899 | WB (1:10,000) |
| Antibody | goat polyclonal anti-mouse IgG Fc (HRP) | abcam | ab97265 RRID:AB_10680426 | WB (1:10,000) |
| Antibody | goat polyclonal anti-rabbit IgG (whole molecule) 10 nm gold preadsorbed | abcam | ab27234 RRID:AB_954427 | TEM (1:50) |
| Antibody | goat polyclonal anti-mouse IgG (whole molecule) 5 nm gold preadsorbed | Sigma/Merck | G7527 RRID:AB_259955 | TEM (1:50) |
| Other | CF488A conjugated Wheat Germ Agglutinin (WGA) | Biotium | 29022-1 | Lot Number: 21C0224-1149057 |
| Chemical compound, drug | Bafilomycin A1 | Sigma/Merck | B1793 | Lot Number: 0000190389 |
| Chemical compound, drug | Colchicine | Serva | 77120.02 | Lot Number: 190300 |
| Chemical compound, drug | Chloroquine diphosphate | Invitrogen | P36236 C | Lot Number: 2441325 |
| Chemical compound, drug | Rapamycin | Sigma/Merck | R0395 | Lot Number: 0000084976 |
| Chemical compound, drug | Cytochalasin B | Sigma | C2743 | Lot Number: 037M4083V |
| Chemical compound, drug | FBS | Biosera | FB-1090/500 | Lot Number: 015BS575 |
| Other | TFF Easy | HansaBioMed Life Sciences | HBM-TFF/1 | |
| Software, algorithm | LASX | Leica | Leica Application Suite X 3.5.5.19976 | |
| Software, algorithm | ZEN Blue | Zeiss | ZEN 2.3 lite | |
| Software, algorithm | iTEM | Olympus | iTEM 5.1 | |
| Software, algorithm | ImageJ | https://imagej.n et/ij/ | v1.54g | |
| Software, algorithm | Prism9 | GraphPad | GraphPad Prism 9.4.1 | |
| Software, algorithm | BioRender | https://www.biorender.com/ |





