Perturbations in eIF3 subunit stoichiometry alter expression of ribosomal proteins and key components of the MAPK signaling pathways
Figures
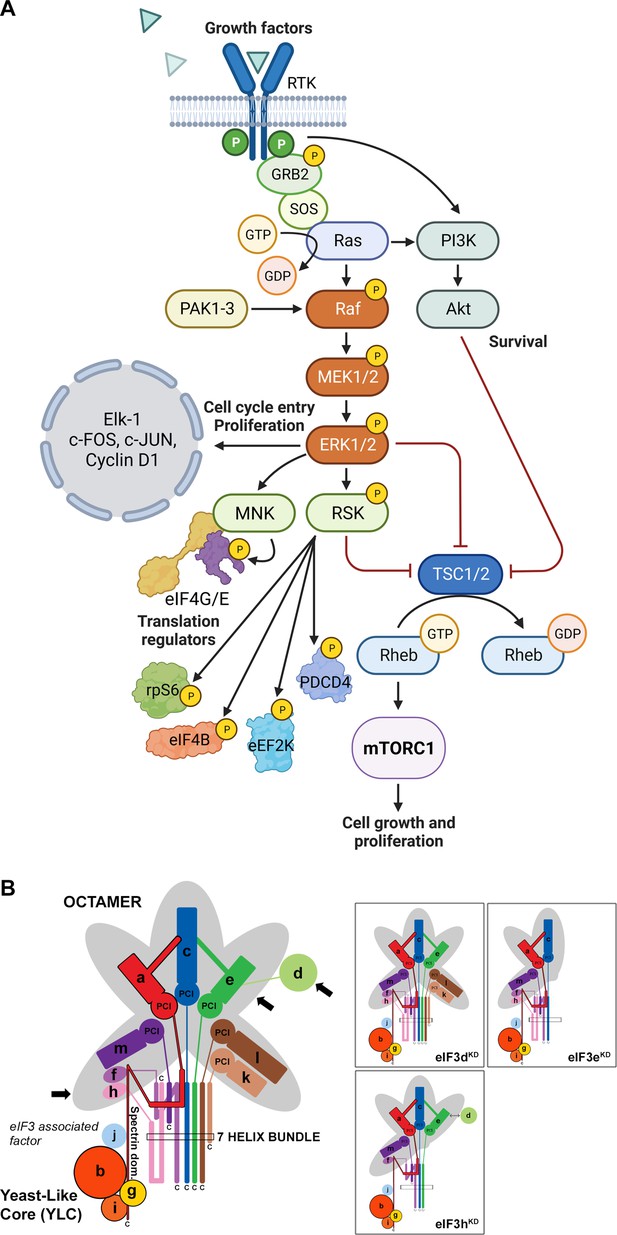
Schematic models of the core of MAPK/ERK pathway and human eIF3 complex.
(A) The MAPK/ERK pathway activation by growth factors and receptor tyrosine kinases (RTKs). RTKs signal through GRB2-Sos and Ras to activate Raf-MEK-ERK signaling cascade. Active ERK phosphorylates numerous substrates, both nuclear and cytoplasmic, to affect gene transcription, protein translation, cell growth, proliferation, and survival. Both RTKs and Ras also activate PI3K-AKT pathway that affect cell survival and growth. Conventional PAKs (PAK1-3) phosphorylate c-Raf and contribute to its activation. (B) eIF3 subunits forming the PCI/MPN octamer are indicated by the grey background. The rectangle marks the seven α-helices involved in formation of the 7-helix bundle. The Yeast-Like Core (YLC) comprising the eIF3 subunits a, b, g, and i is depicted, and so is the eIF3-associated factor eIF3j. Arrows indicate subunits targeted by siRNA and subjected to Ribo-Seq in this study; eIF3 subcomplexes generated in individual knock-downs are boxed (adapted from Figure 6 of Wagner et al., 2016). This figure was created with BioRender.com.
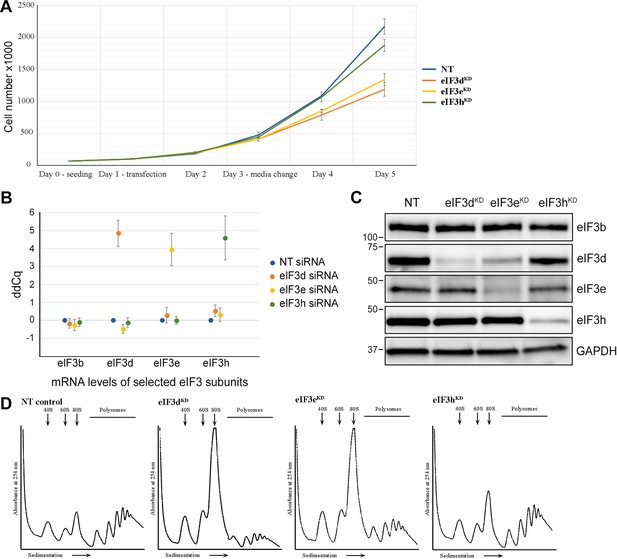
Quality control of eIF3e, d, and h knock-downs - Hela cells downregulated for eIF3d, eIF3e, and eIF3h display slower growth and impaired translation.
(A) Growth curve of Hela cells transfected with siRNA targeting eIF3d, eIF3e, eIF3h or non-targeting (NT) siRNA as a control. Cells were grown in 6 well plates in duplicates and counted by Corning cell counter. (B) mRNA levels of the selected downregulated eIF3 subunits were significantly decreased as assessed by quantitative PCR for all replicates. Plot represents the result of all replicates used for RNA-Seq and Ribo-Seq library preparations, mean ± SD. The ddCq value displays the threshold cycle normalized to the reference gene ALAS1 and to control cells transfected with non-targeting siRNA (NT). The ddCq values are in log2 scale. A ddCq of 0 indicates no change to NT siRNA control; ddCq = 4 indicates a drop to 6.25% (16 fold) compared to NT siRNA control. (C) Downregulation of selected eIF3 subunits targeted by siRNA was determined by Western blotting with GAPDH used as a loading control. (D) Standard polysome profiles illustrate that cells were not overgrown at the time of harvest and occurred in a state of active translation (NT) with the expected specific reduction in polysomes in all three knock-downs, as reported previously (Wagner et al., 2016). One example profile for each knock-down is shown.
-
Figure 1—figure supplement 1—source data 1
Original files for western blot analysis displayed in Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
PDF file containing original western blots for Figure 1—figure supplement 1C, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig1-figsupp1-data2-v1.pdf
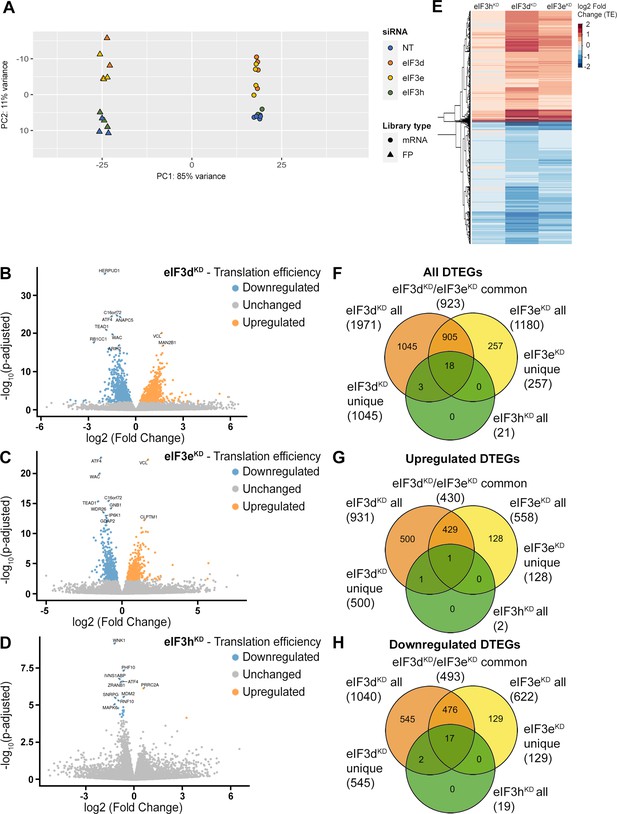
DTEGs identified in eIF3dKD, eIF3eKD, and eIF3hKD largely overlap.
(A) Principal component analysis of read count per gene of the RNA-Seq and Ribo-Seq libraries. (B - D) Volcano plots of significant (Padjusted <0.05) DTEGs in individual knock-downs: eIF3dKD (B), eIF3eKD (C) and eIF3hKD (D). The volcano plots show the Log2 fold-change of TE (x-axis) versus the significance -Log10 p-adjusted value (y-axis). Genes without significant p-adjusted value are plotted in grey, downregulated DTEGs are plotted in blue and upregulated DTEGs in orange. For each plot, the top 10 most significant DTEGs are labeled with gene names. The Volcano plots were generated using modified script from Galaxy (Blankenberg et al., 2014). (E) Heatmap and dendrogram resulting from hierarchical clustering analysis of significant TE changes observed in eIF3dKD, eIF3eKD, and eIF3hKD cells. (F - H) Venn diagrams depicting the overlap in all DTEGs (F), in upregulated DTEGs (G), and in downregulated DTEGs (H) among eIF3dKD, eIF3eKD, and eIF3hKD. Based on the overlaps, DTEGs were divided in several subgroups for further analysis: eIF3dKD ‘all’, eIF3dKD ‘unique’, eIF3eKD ‘all’, eIF3eKD ‘unique’, eIF3dKD/eIF3eKD ‘common’ and eIF3hKD ‘all’.
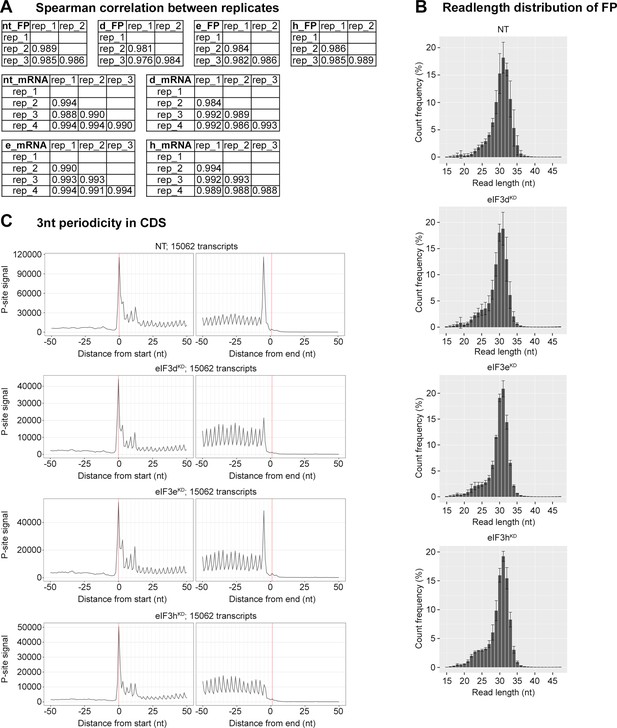
Quality control of Ribo-Seq libraries from all eIF3 knock-downs and the NT control.
(A) Spearman correlation coefficients of the footprint or mRNA count per gene among all replicates. (B) Read length distribution of footprints in Ribo-Seq libraries from all eIF3 knock-downs and the NT control. Bar charts show average values for each fragment length (after alignment to genome); all Ribo-Seq libraries were done in triplicates. (C) Meta-profiles showing the periodicity of ribosomes along the transcripts at the genome-wide scale. The metaprofiles are based on the P-site identification obtained by using riboWaltz (Lauria et al., 2018).
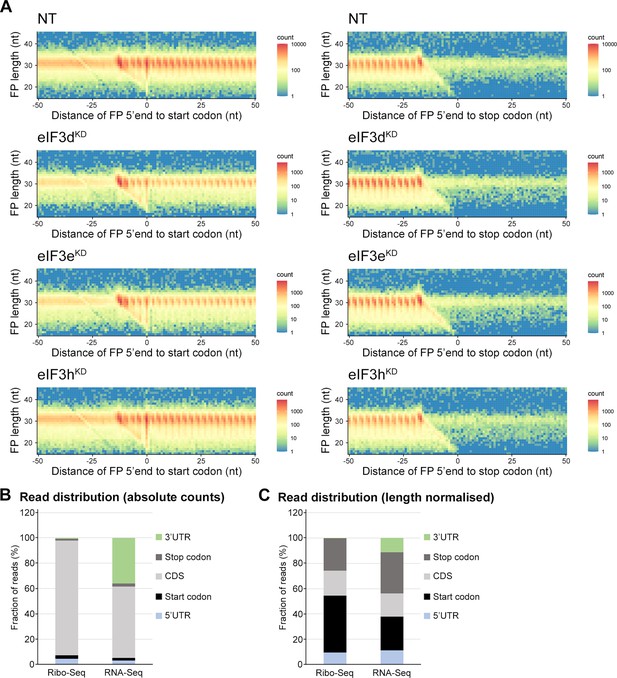
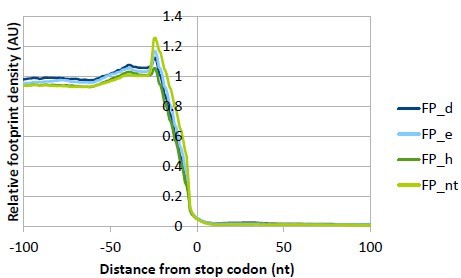
Ribo-Seq libraries from all eIF3 knock-downs and the NT control display triplet periodicity and enrichment of reads in CDS.
(A) All Ribo-Seq libraries show typical 3nt periodicity in the CDS. Metagene plots of footprint (FP) length versus 5’ end position relative to the first nucleotide (position 0) of start (left) or stop codons (right). The color scale represents FP count as indicated on the right and is plotted in log scale. The labels at the color bar are given in linear scale. Heatmaps are shown for one exemplary replicate from each sample. (B) Fraction of reads in different transcript features (C) Same as in (B) but numbers of reads in each feature were normalized for average feature length.
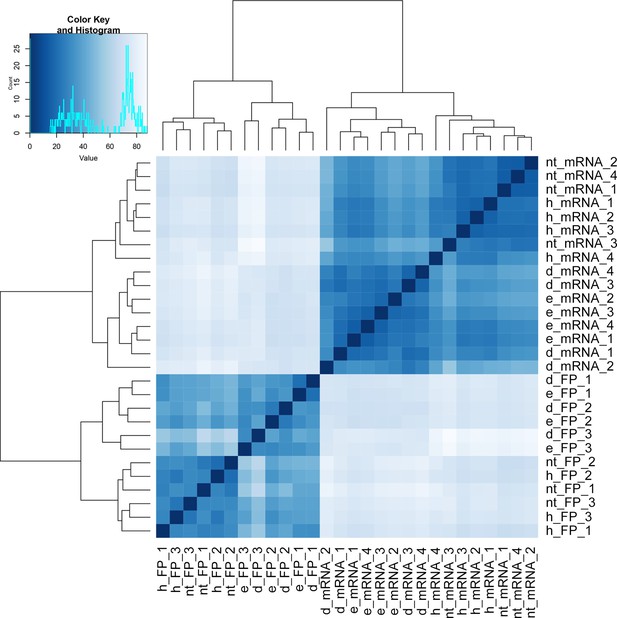
Clustering of Ribo-Seq and RNA-Seq libraries.
Heatmap and dendrogram resulting from hierarchical clustering analysis of Ribo-Seq libraries and RNA-Seq libraries. Analysis was performed on gene counts by DESeq2 (Love et al., 2014) and biomaRt (Durinck et al., 2009).
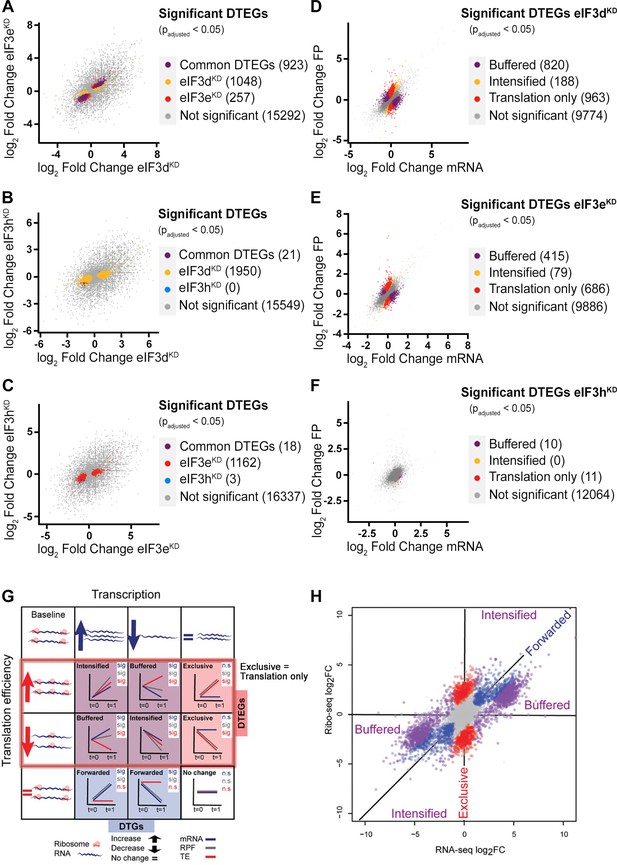
DTEGs identified in all knock-downs largely overlap.
(A–C) Scatter plots of genes with significant TE changes and their overlap in individual knock-downs. (A) eIF3dKD vs. eIF3eKD (B) eIF3dKD vs. eIF3hKD (C) eIF3eKD vs. eIF3hKD. (D–F) Scatter plots showing classification of significant DTEGs into different groups based on fold changes of FP and mRNA. Buffered DTEGs (purple) display a significant change in TE that counteracts the change in mRNA, hence buffering the effect of transcription. Translation only DTEGs (red) have a significant change in FP while maintaining the same mRNA levels, which results in a significant change in TE. Translationally intensified DTEGs (yellow) have a significant change in TE that occurs in the same direction as the effect of transcription (i.e. a gene exhibiting an increase in transcription also exhibits increase in TE that altogether boost protein production). (D) eIF3dKD, (E) eIF3eKD, (F) eIF3hKD. (G, H) Classification of genes based on fold changes of FP (RFP), mRNA, and TE (reproduced from Figure 1 of Chothani et al., 2019). (G) A gene could be either DTG (Differentially Transcribed Gene) and/or DTEG (Differential Translation Efficiency Gene), and based on the direction of change would fall into one of the eight gene-regulatory possibilities (sig: significant, n.s.: not significant). Translationally forwarded genes are DTGs that have a significant change in mRNA and FP at the same rate, with no significant change in TE. Conversely, translationally exclusive / translation only genes are DTEGs that have a significant change in FP, with no change in mRNA leading to a significant change in TE. Several genes are both DTGs and DTEGs, and their regulatory class is determined based on a combination of the relative direction of change between transcription and translation efficiency. Specifically, translationally buffered genes have a significant change in TE that counteracts the change in RNA; hence, buffering the effect of transcription. Translationally intensified genes have a significant change in TE that acts with the effect of transcription. In all cases, the change in RNA can be either positive or negative, and where buffering or intensifying takes place, the direction of change is taken into account. For example, a gene that exhibits an increase in transcription and an increase in translation efficiency is classified as intensified, while a gene that exhibits an increase in transcription but a decrease in translational efficiency is classified as buffered. (H) Simulated data showing fold changes for each gene in RNA-seq and Ribo-seq data. Translationally forwarded genes (in blue), exclusive (translation only) genes (in red), buffered genes (in purple), and intensified genes (in purple) are highlighted.
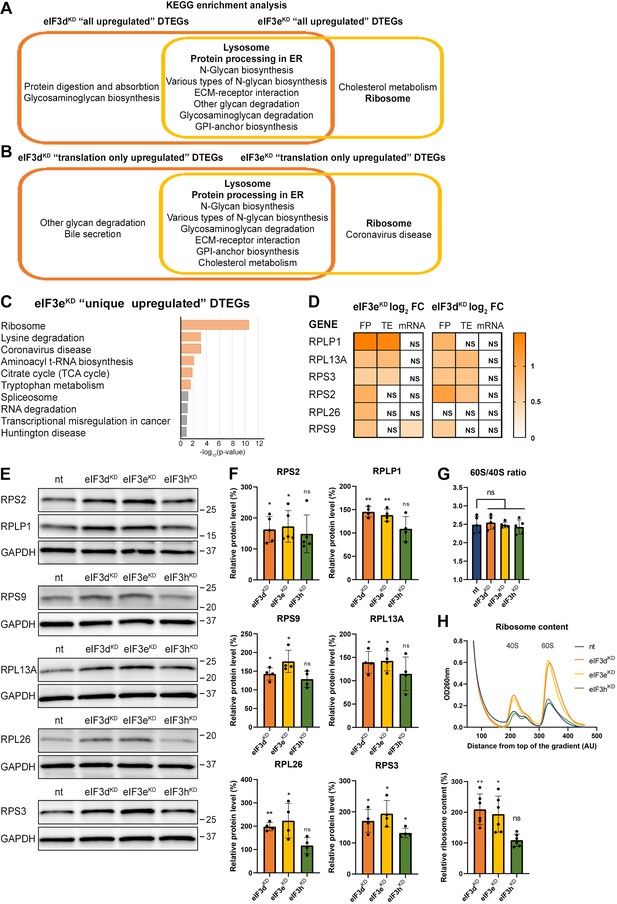
KEGG pathway enrichment analysis of the eIF3dKD- and eIF3eKD-associated upregulated DTEGs reveals upregulation of ribosomal proteins.
(A) Venn diagram of the 10 most significantly enriched KEGG pathways for eIF3dKD ‘all’ and eIF3eKD ‘all’ groups of upregulated DTEGs, highlighting that most of the pathways are common to both knock-downs. Lysosome and Protein processing in ER are highlighted in bold. The complete results of the KEGG enrichment analysis with corresponding p-values can be found in Figure 3—figure supplement 1A. (B) Venn diagram as in A but displaying eIF3dKD ‘translation only’ and eIF3eKD ‘translation only’ groups of the upregulated DTEGs. (C) The bar chart shows the top 10 enriched KEGG terms for eIF3eKD ‘unique’ upregulated DTEGs. Orange bars correspond to terms with significant p-values (<0.05), grey bars correspond to terms with not significant p-values (>0.05). The p-values were calculated by Enrichr gene set search engine. (D) The list of genes pre-selected for western blot analysis and a heatmap showing their respective log2 fold-change values from differential expression analysis of FP, TE and mRNA in eIF3eKD and eIF3dKD. Positive values indicating significant upregulation are in shades of orange. ns = not significant p-adjusted value. (E) Western blot analysis of selected ribosomal proteins performed in the indicated knock-downs and the NT control. GAPDH was used as a loading control. (F) Relative protein levels of selected ribosomal proteins normalized to GAPDH; plots show mean ± SD, NT control = 100%. Dots represent results from individual biological replicates. Shapiro-Wilk test was used to test for normal distribution. One sample t-test was used for statistical evaluation, p-values: *=p < 0.05, **=p < 0.01, ns = not significant. (G) eIF3dKD, eIF3eKD, and eIF3hKD do not influence the balanced ribosomal subunits production. The 60 S/40 S ratio was calculated from polysome profiles carried out in the presence of 50 mM EDTA. Dots represent results from individual biological replicates. Plots show mean ± SD. Shapiro-Wilk test was used to test for normal distribution. Paired t test was used for statistical evaluation, all downregulations were individually compared to NT. ns = not significant p-value. (H) Ribosomal content is increased in eIF3dKD and eIF3eKD. One representative polysome profile, carried out in the presence of 50 mM EDTA, made from 10 million cells is shown in the upper panel. Relative ribosomal content normalized to NT control set to 100% is shown in the lower panel. Plot shows mean ± SD. Individual biological replicates are depicted as dots. Shapiro-Wilk test was used to test for normal distribution. One sample t-test was used for statistical evaluation, p-values: *=p < 0.05, **=p < 0.01. All plots in (F–H) were created in GraphPad Prism version 8.4.3 for Windows.
-
Figure 3—source data 1
Original files for western blot analysis displayed in Figure 3E.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig3-data1-v1.zip
-
Figure 3—source data 2
PDF file containing original western blots for Figure 3E, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig3-data2-v1.pdf
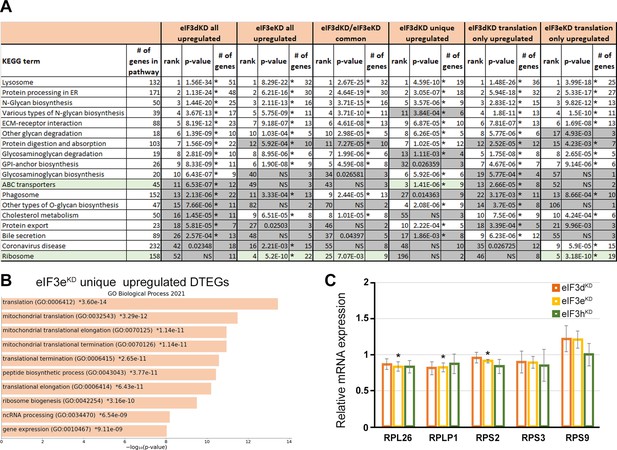
KEGG pathway enrichment analysis of the eIF3dKD- and eIF3eKD-associated upregulated DTEGs reveals upregulation of ribosomal proteins.
(A) List of top 10 significantly enriched KEGG pathways for eIF3dKD ‘all’, eIF3eKD ‘all’, eIF3dKD/eIF3eKD ‘common’, eIF3dKD ‘unique’, eIF3dKD ‘translation only’, and eIF3eKD ‘translation only’ groups of upregulated DTEGs. Ranking corresponds to the lowest-to-highest p-value (calculated by Enrichr gene set search engine), with the ‘eIF3dKD all’ group setting the primary ranking. Less significant terms ranking 11 and below are in grey. Terms specifically discussed in the main text are highlighted in green. For each term, the total number of genes in the pathway and number of DTEGs found in each pathway is indicated. Only significant p-values (<0.05) are shown. NS = not significant p-value (>0.05) Asterisk next to a p-value indicates that a given term also has a significant Benjamini-Hochberg adjusted p-value (<0.05). (B) The bar chart shows the top 10 enriched GO Biological Process terms for eIF3eKD ‘unique upregulated’ DTEGs, along with their corresponding p-values. (C) Relative mRNA expression of selected Ribosomal protein genes normalized to NT control = 1. One-sample t-test was used for statistical evaluation, *=p < 0.05, N=3.
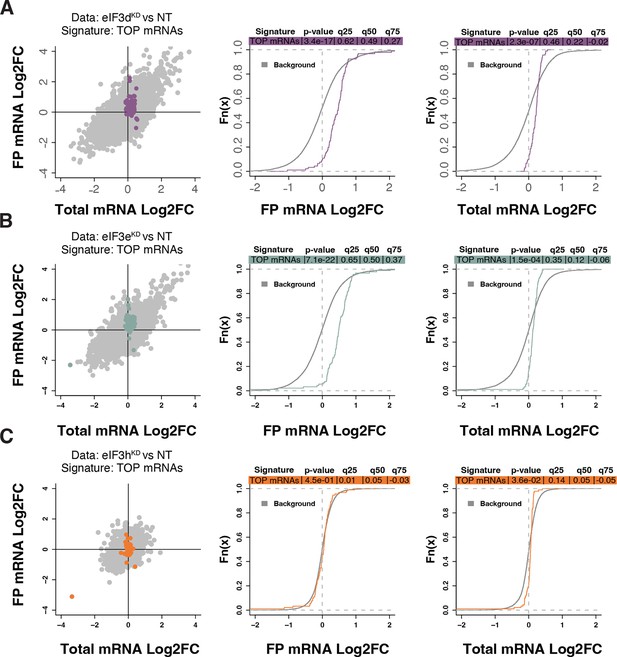
Loss of eIF3 subunits leads to translational activation of mRNAs with 5’UTR TOP motifs.
(A–C) Scatterplots from translatome analysis of eIF3dKD (A), eIF3eKD (B), and eIF3hKD (C) with the location of transcripts harboring 5’ UTR TOP motifs (Philippe et al., 2020) colored (left panels). Middle and right panels show the empirical cumulative distribution functions of log2 fold changes in FP and total mRNA for the transcripts with TOP motifs. The background constituting all other transcripts are shown as grey curves. Significant differences between the distributions were identified using the Wilcoxon rank-sum test. Differences between the distributions at each quantile are indicated. Shift to the right indicates increased expression, while shift to the left indicates decreased expression. eIF3dKD and eIF3eKD show very significant increase of FPs of TOP mRNAs, suggesting mainly translational upregulation.
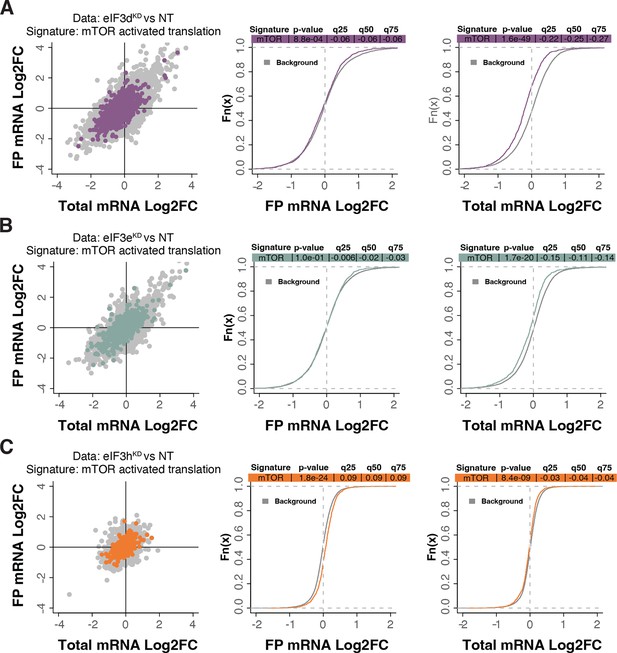
mTOR-sensitive transcripts tend to be translationally offset (buffered) with loss of eIF3d and eIF3e.
(A–C) Scatterplots from translatome analysis of eIF3dKD (A), eIF3eKD (B), and eIF3hKD (C) with the location of transcripts that are translationally activated by insulin stimulation (mTOR activation) (Gandin et al., 2016) colored (left panels). Middle and right panels show the empirical cumulative distribution functions of log2 fold changes in FP and total mRNA for the transcripts that are translationally activated by mTOR signaling. The background constituting all other transcripts are shown as grey curves. Significant differences between the distributions were identified using the Wilcoxon rank-sum test. Differences between the distributions at each quantile are indicated. mRNAs of mTOR-sensitive transcripts in eIF3dKD and eIF3eKD display significant downregulation (shift to the left) while the FPs remain unchanged, suggesting translational buffering.
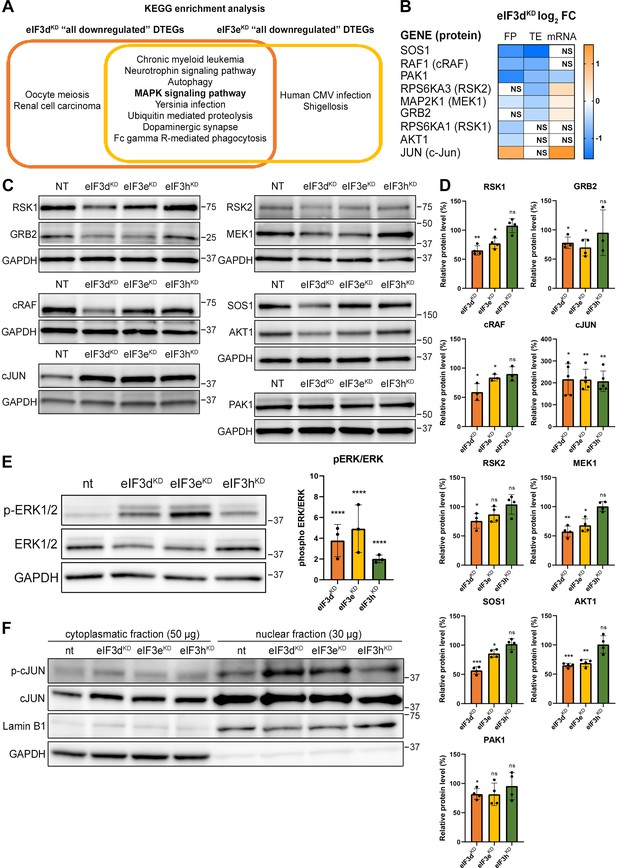
KEGG pathway enrichment analysis of the eIF3dKD- and eIF3eKD-associated downregulated DTEGs reveals downregulation of MAPK signaling pathways components.
(A) Venn diagram of the 10 most significantly enriched KEGG pathways for eIF3dKD ‘all’ and eIF3eKD ‘all’ groups of downregulated DTEGs, highlighting that most of the pathways are common to both knock-downs. The MAPK signaling pathway is highlighted in bold. The complete results of the KEGG enrichment analysis with corresponding p-values can be found in Figure 5—figure supplement 1A. (B) The list of genes pre-selected for western blot analysis and a heatmap showing their respective log2 fold-change values from differential expression analysis of FP, TE and mRNA in eIF3dKD. Negative values indicating a significant downregulation are in shades of blue, while positive values showing significant upregulation are in shades of orange. ns = not significant p-adjusted value. (C) Western blot analysis of selected proteins constituting the MAPK/ERK pathway performed in the indicated knock-downs and the NT control. GAPDH was used as a loading control. (D) Relative protein levels of selected MAPK/ERK pathway proteins normalized to GAPDH; plots show mean ± SD, NT control = 100%. Dots represent results from individual biological replicates. Shapiro-Wilk test was used to test for normal distribution. One sample t-test was used for statistical evaluation, p-values: *=p < 0.05, **=p < 0.01, ***=p < 0.001, ns = not significant. (E) Western blot analysis of the phosphorylation status of the ERK1/2 proteins (left) and its relative quantification normalized to NT = 1 (right). Plot shows mean ± SD. Dots represent results from individual biological replicates. Shapiro-Wilk test was used to test for normal distribution. One-sample t-test was used for statistical evaluation, ****=p < 0.0001. (F) Western blot analysis of the phosphorylation status of the c-Jun transcription factor in cytoplasmatic and nuclear fractions in the indicated knock-downs and the NT control. The protein loading was 50 µg of total protein for cytoplasmatic lysate and 30 µg of total protein for nuclear lysate, as indicated. GAPDH was used as cytoplasmatic loading control and Lamin B1 was used as nuclear loading control. This experiment was repeated three times with similar results. All plots in (D, E) were created in GraphPad Prism version 8.4.3 for Windows.
-
Figure 5—source data 1
Original files for western blot analysis displayed in Figure 5C, E and F.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig5-data1-v1.zip
-
Figure 5—source data 2
PDF file containing original western blots for Figure 5C, E and F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig5-data2-v1.pdf
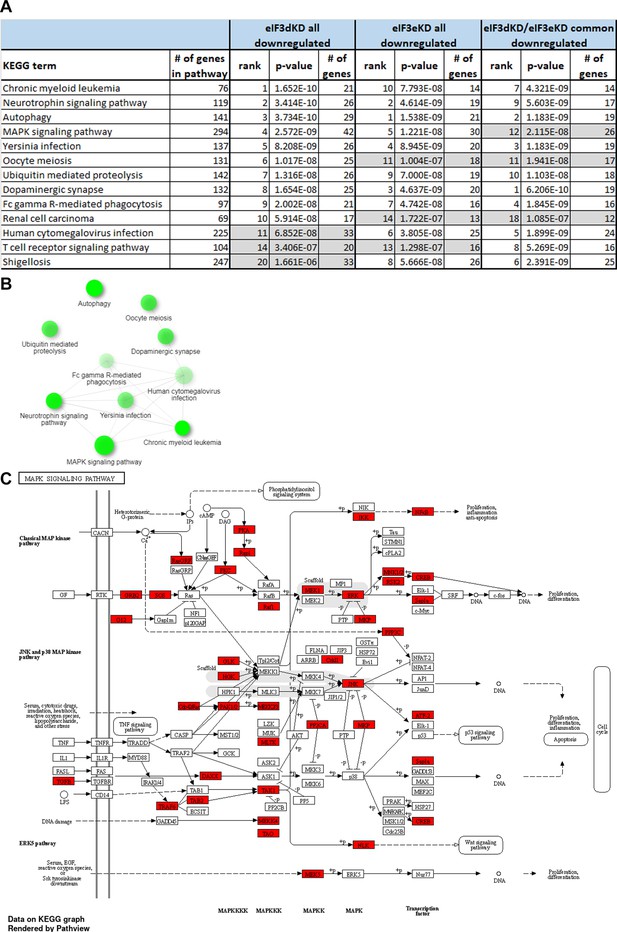
KEGG pathway enrichment analysis of the eIF3dKD- and eIF3eKD-associated downregulated DTEGs reveals downregulation of ‘MAPK signaling pathway’ components.
(A) List of top 10 significantly enriched KEGG pathways for eIF3dKD ‘all’, eIF3eKD ‘all’, and eIF3dKD/eIF3eKD ‘common’ groups of downregulated DTEGs. Ranking corresponds to the lowest-to-highest p-value (calculated by Enrichr gene set search engine), with the eIF3dKD ‘all’ group setting the primary ranking. Less significant terms ranking 11 and below are in grey. For each term, the total number of genes in the pathway and number of DTEGs found in each pathway is indicated. All results presented have also significant adjusted p-values as calculated using the Benjamini-Hochberg (BH) procedure to account for multiple hypotheses. (B) Network graph of the top 10 most significantly enriched KEGG terms for the eIF3dKD ‘all downregulated’ group of DTEGs. Related KEGG terms are connected with a solid line if they share 20% or more genes. Darker nodes represent more significantly enriched gene sets. Greater nodes represent larger gene sets. (C) The KEGG MAPK signaling pathway hsa04010 scheme. Downregulated DTEGs in eIF3dKD matching the genes of this pathway are highlighted in red. See text and Supplementary file 1 for further details.
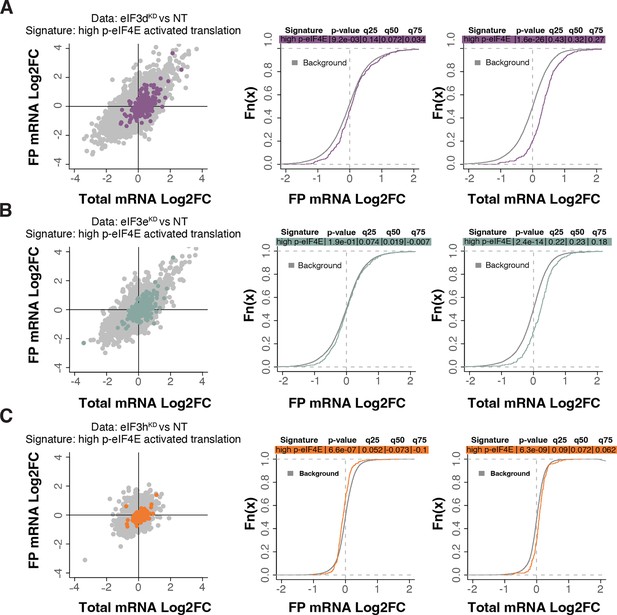
Loss of eIF3 subunits leads to translational offsetting of transcripts with enhanced translation downstream of phosphorylated eIF4E.
(A–C) Scatterplots from translatome analysis of eIF3dKD (A), eIF3eKD (B), and eIF3hKD (C) with the location of transcripts translationally activated by high phosphorylation of eIF4E (Karampelias et al., 2022) colored (left panels). Middle and right panels show the empirical cumulative distribution functions of log2 fold changes in FP and total mRNA for the p-eIF4E sensitive transcripts. The background constituting all other transcripts are shown as grey curves. Significant differences between the distributions were identified using the Wilcoxon rank-sum test. Differences between the distributions at each quantile are indicated. mRNAs of phospho-eIF4E sensitive transcripts in eIF3dKD and eIF3eKD display significant upregulation (shift to the right) while the FPs remain unchanged, suggesting translational buffering.
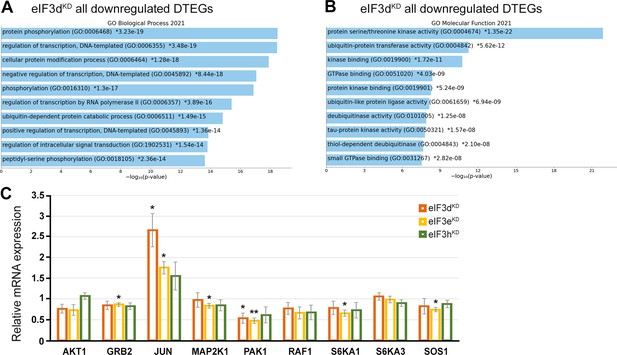
GO enrichment analysis for eIF3dKD associated downregulated DTEGs.
(A) The bar chart shows the top 10 enriched GO Biological Process terms for eIF3dKD ‘all downregulated’ DTEGs, along with their corresponding p-values. (B) The bar chart shows the top 10 enriched GO Molecular Function terms for eIF3dKD ‘all downregulated’ DTEGs, along with their corresponding p-values. (A, B) Colored bars correspond to terms with significant p-values (<0.05). Asterisk indicates that a given term also has a significant adjusted p-value (<0.05). All p-values were calculated by Enrichr gene set search engine. (C) Relative mRNA levels of selected genes from MAPK/ERK signaling pathway normalized to NT control = 1. One-sample t-test was used for statistical evaluation, p-values: *=p < 0.05, **=p < 0.01, N=3.
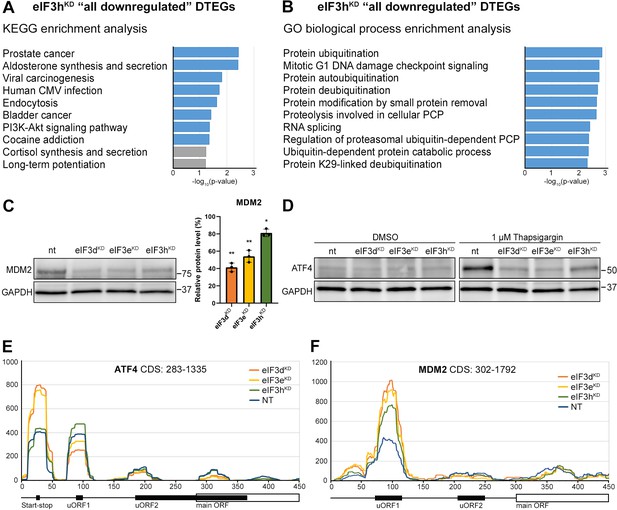
KEGG pathway and GO enrichment analysis of the eIF3hKD-associated DTEGs reveals downregulation of proto-oncogene MDM2 and a defective stress-induced upregulation of ATF4.
(A) The bar chart shows the top 10 enriched KEGG terms for eIF3hKD ‘all downregulated’ DTEGs. Blue bars correspond to terms with significant p-values (<0.05), grey bars correspond to terms with not significant p-values (>0.05). The p-values were calculated by Enrichr gene set search engine. (B) The bar chart shows the top 10 enriched GO Biological Process terms for eIF3hKD ‘all downregulated’ DTEGs. Blue bars correspond to terms with significant p-values (<0.05). The p-values were calculated by Enrichr gene set search engine. PCP; protein catabolic process. (C) Western blot analysis of the MDM2 expression preformed in the indicated knock-downs and NT control. GAPDH was used as a loading control; plot shows mean ± SD, NT control = 100%. Dots represent results from individual biological replicates. Shapiro-Wilk test was used to test for normal distribution. One sample t-test was used for statistical evaluation, p-values: *=p < 0.05, **=p < 0.01. Plots was created in GraphPad Prism version 8.4.3 for Windows. (D) Western blot analysis of the stress-induced upregulation of ATF4 expression performed in the indicated knock-downs and NT control. Before harvesting, cells were incubated for 3 hr with 1 µM Thapsigargin to induce ER stress or with DMSO as a stress-free control. GAPDH was used as a loading control. (E) Normalized ribosomal footprint coverage along the ATF4 mRNA (first 450 nucleotides). Schematic with regulatory elements is shown at the bottom. Average of all three replicates is shown. Footprint coverage was normalized to all footprints mapping to the ATF4 mRNA. (F) Same as in (E) only for MDM2 mRNA.
-
Figure 6—source data 1
Original files for western blot analysis displayed in Figure 6C and D.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig6-data1-v1.zip
-
Figure 6—source data 2
PDF file containing original western blots for Figure 6C and D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/95846/elife-95846-fig6-data2-v1.pdf
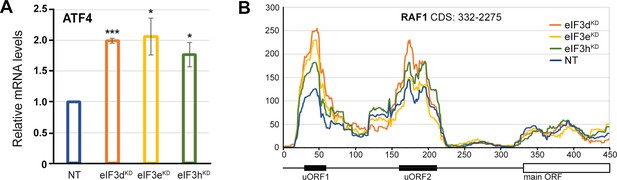
The ATF4 mRNA is upregulated in all three eIF3 knock-downs tested.
(A) Relative mRNA levels of ATF4 transcription factor. One-sample t-test was used for statistical evaluation, *=p < 0.05, ***=p < 0.001, N=3. (B) Normalized ribosomal footprint coverage along the RAF1 mRNA (first 450 nucleotides). Schematic with regulatory elements is shown at the bottom. Average of all three replicates is shown. Footprint coverage was normalized to all footprints mapping to RAF1 mRNA.
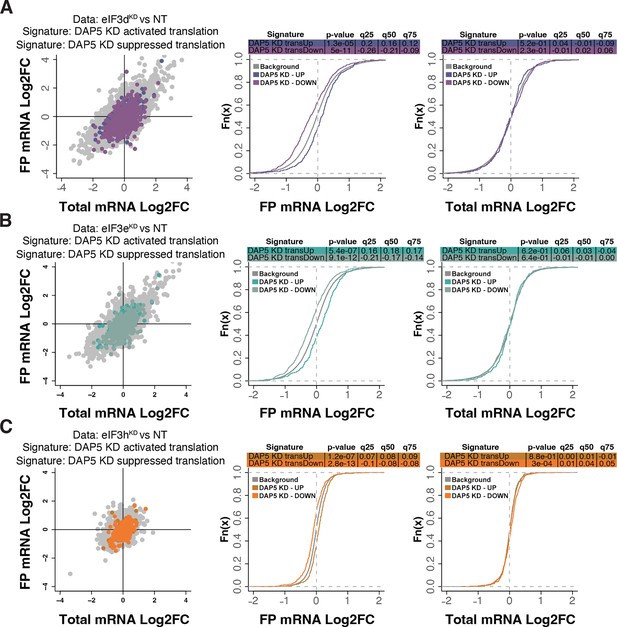
Loss of eIF3 subunits modulates signatures of DAP5-dependent translation.
(A–C) Scatterplots from translatome analysis of eIF3dKD (A), eIF3eKD (B), and eIF3hKD (C) with the location of transcripts that are translationally activated and suppressed by KD of DAP5 (David et al., 2022) colored (left panels). Middle and right panels show the empirical cumulative distribution functions of log2 fold changes in FP and total mRNA for the transcripts that are translationally activated or suppressed upon loss of DAP5. The background constituting all other transcripts are shown as grey curves. Significant differences between the distributions were identified using the Wilcoxon rank-sum test. Differences between the distributions at each quantile are indicated.
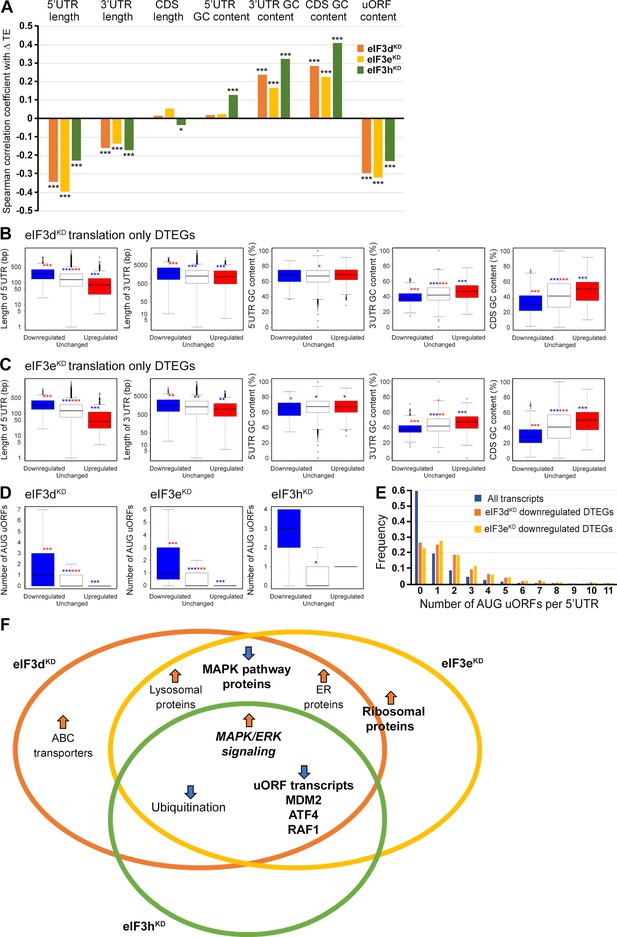
Differential TE transcripts in eIF3dKD and eIF3eKD show negative correlation with the UTR length and uORF content while they positively correlate with GC content of their 3’ UTRs and coding sequences (CDS).
(A) Bar plot showing the Spearman correlation between the observed ΔTE values for all genes with assigned adjusted p-values in each knock-down and different mRNA features (5’ and 3’ UTR length or GC content, uORF content in 5’ UTR). ***=p < 10–20, **=p < 10–10, *=p < 0.005. (B, C) Box and whisker plots comparing UTR lengths or GC content (in UTRs or CDS) among mRNAs with TE significantly increased (red, upregulated), decreased (blue, downregulated), or unchanged (white) in eIF3dKD (B) and eIF3eKD (C); ***=Padj < 10–10, **=Padj < 10–5, *=Padj < 0.05, color indicates comparison set. (D) Same as in (B, C) but for the number of AUG-initiated uORFs in 5’ UTRs in the eIF3d, eIF3e, and eIF3h knock-downs. Outliers are not shown for better clarity. (E) Histogram of the frequency of AUG-initiated uORFs per 5’ UTR in all transcripts listed in the uORFdb database that were assigned a p-value in this study (n=11450), for downregulated translation only DTEGs in eIF3dKD (n=1027) and for downregulated translation only DTEGs in eIF3eKD (n=618). (F) Venn diagram summarizing the main results of this study. Groups of significant DTEGs identified in given knock-downs (encircled – color-coded) are indicated by a blue arrow for downregulated or an orange arrow for upregulated. Results confirmed by western blotting are shown in bold. Ribosomal proteins were placed on the borderline between eIF3dKD and eIF3eKD because they were identified as eIF3eKD ‘unique upregulated’ DTEGs, but their protein levels were elevated in both eIF3eKD and eIF3dKD, as shown by western blotting. The ‘MAPK/ERK signaling’ is shown in bold italics because it is an independent phenomenon that could be detected only by western blotting.
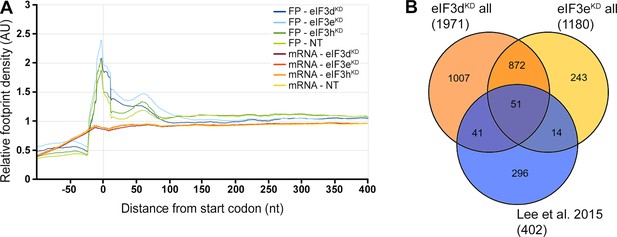
eIF3dKD, eIF3eKD, and eIF3hKD do not show accumulation of footprints at the beginning of CDSes, as reported previously, but display a significant overlap with mRNAs directly interacting with eIF3.
(A) Metagene plot of ribosome density distribution around the start codon of all mRNAs in eIF3dKD, eIF3eKD, and eIF3hKD. The plot shows averages of triplicates from Ribo-Seq and quadruplicates from RNA-Seq. (B) Venn diagram of the overlap between eIF3d/eKD DTEGs and mRNAs directly interacting with eIF3 as determined by the CLIP assay in Lee et al., 2015.
Additional files
-
Supplementary file 1
Complete differential expression data.
- https://cdn.elifesciences.org/articles/95846/elife-95846-supp1-v1.xlsx
-
Supplementary file 2
KEGG pathway and GO Biological Process enrichment analysis of eIF3hKD downregulated DTEGs.
- https://cdn.elifesciences.org/articles/95846/elife-95846-supp2-v1.docx
-
Supplementary file 3
Material tables – siRNAs, qPCR primers and antibodies used in this study.
- https://cdn.elifesciences.org/articles/95846/elife-95846-supp3-v1.docx
-
Supplementary file 4
Statistics of read processing for all Ribo-Seq and RNA-Seq libraries.
- https://cdn.elifesciences.org/articles/95846/elife-95846-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95846/elife-95846-mdarchecklist1-v1.docx