Transcription factor condensates, 3D clustering, and gene expression enhancement of the MET regulon
Figures
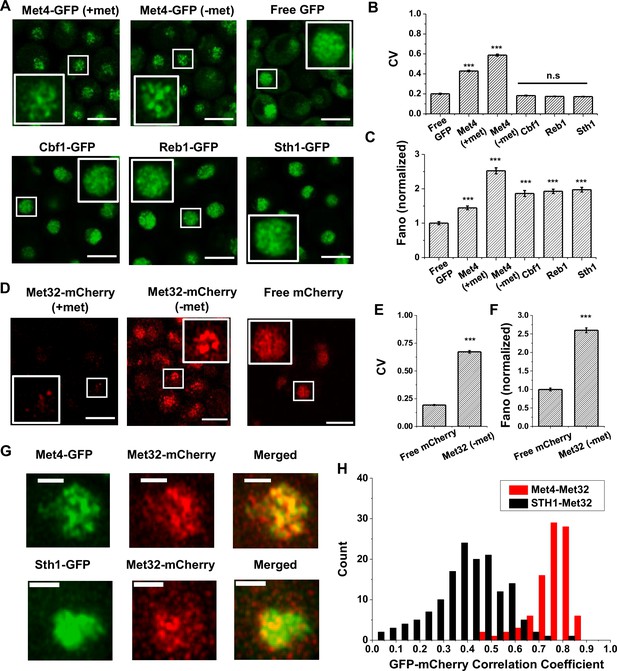
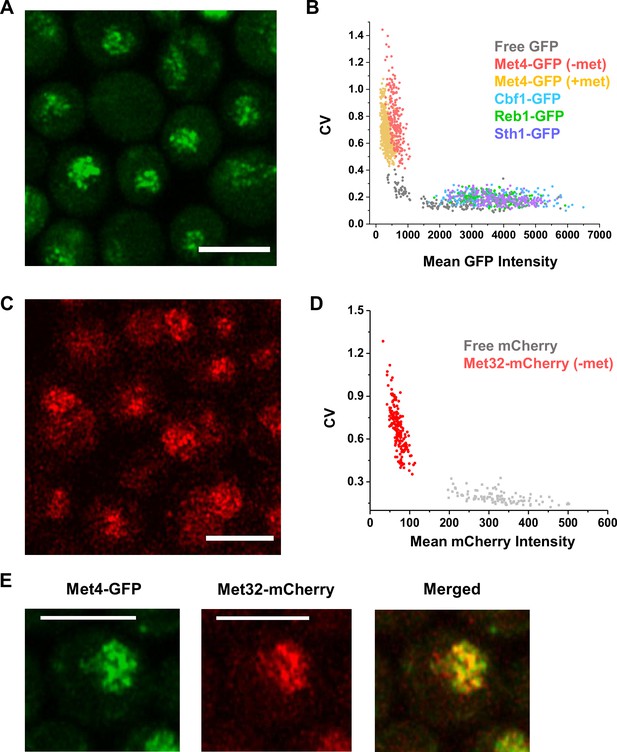
The trans-activator Met4 and its co-factor Met32 form puncta in the nucleus.
(A) Representative images of Met4-GFP in ± methionine (met) conditions. Control cells expressing free GFP (driven by the HOpr), Cbf1-GFP, Reb1-GFP, and Sth1-GFP cells are shown with similar contrasts. Met4-GFP in −met was imaged four times, and all the other strains/conditions twice. Scale bars represent 4 μm (same as in D and G). (B, C) The mean coefficient of variance (CV) and Fano numbers of nuclear pixel intensities for different versions of GFPs in panel A. The Fano numbers are normalized so that the free GFP has a Fano number 1. Error bar represents standard error among all cells. Significance was calculated in comparison with free GFP values using two-tailed Student’s t-test (***p < 0.001). Number of cells analyzed: Met4-GFP +/−met (65/147), free GFP (67), Cbf1 (48), Reb1 (50), Sth1 (59). (D) Representative images of strains expressing Met32-mCherry in +/−met conditions. Control strain expressing free mCherry (driven by the HOpr) is shown with similar contrasts. All strains/conditions were imaged three times. (E, F) The mean CV and normalized Fano number of pixel intensities of Met32-mCherry vs free mCherry. Same statistical test was used as in B and C. (G) Representative images of strains co-expressing Met4-GFP and Met32-mCherry (top row), or Sth1-GFP and Met32-mCherry (bottom row). Both strains were imaged three times. (H) A histogram of Pearson correlation coefficient between co-expressed Met4-GFP and Met32-mCherry signals (red bars N = 93) or Sth1-GFP and Met32-mCherry signals (black bars N = 149) in each cell.
High resolution imaging of endogenously labeled Met4 and Met32.
(A) Additional images of Met4-GFP in live yeast cells under −met condition. Scale bars represent 4 µm. Same for below. (B) Coefficient of variance (CV) of pixel intensities vs average intensity in cells expressing various GFP fusions as in Figure 1A. Each dot represents a single nucleus. CV decreases with increasing GFP concentration, and different concentrations of different factors are taken into account by the Fano number. (C) Additional images of Met32-mCherry in live yeast cells under −met condition. (D) Same as in B for free mCherry and Met32-mCherry. (E) Additional images of Met4-GFP and Met32-mCherry co-expressed in live yeast cells under −met condition.
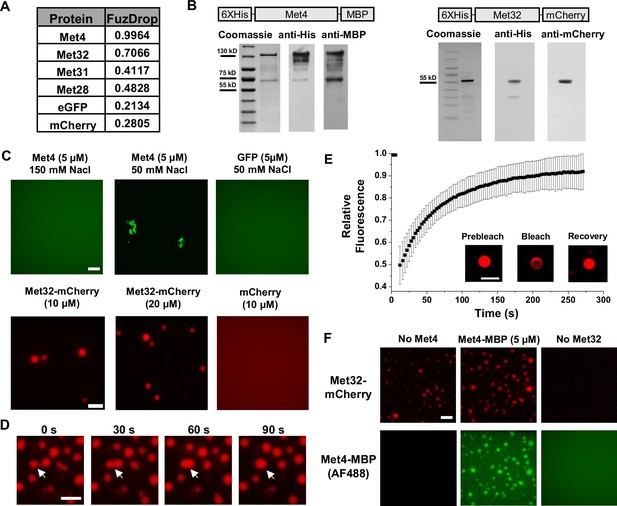
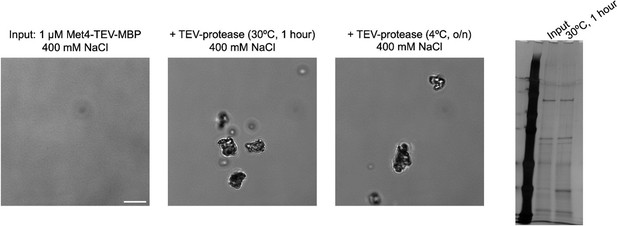
Met32 forms condensates with liquid–liquid phase separation (LLPS) properties in vitro.
(A) Probabilities of individual Met transcription factors (TFs) (Met4, Met32, Met31, Met28), GFP, and mCherry to undergo LLPS from a published prediction program (Fuzdrop). (B) Coomassie staining and western blots of purified Met4 and Met32 proteins. For each protein, three purifications were performed and individually imaged. (C) Protein aggregate and droplet formation observed for Met4-MBP and Met32-mCherry fusion proteins. Met4 and GFP are 5 µM, Met32-mCherry and mCherry are 10 µM in 20 mM HEPES (2-(4-(2-hydroxyethyl)piperazin-1-yl)ethanesulfonic acid) pH 7.5, 150 mM NaCl buffer. Met4 and GFP are in 20 mM HEPES pH 7.5, 150 mM NaCl or 50 mM NaCl. Met4 is labeled with Alexa 488 (AF488). Scale bar represents 10 µm (same as in D–F). (D) Merging of Met32-mCherry droplets. Met32-mCherry is at 30 µM in 20 mM HEPES pH 7.5, 150 mM NaCl. (E) Fluorescence recovery after photobleaching (FRAP) data for 20 µM Met32 droplets. The intensity data was collected every 3 s for 270 s and normalized to percent bleaching. Error bars represent the standard deviation of three biological replicates. Inset: Representative images of Met32 FRAP. (F) Co-localization of Met32-mCherry droplets (10 µM) with Met4-MBP (5 µM) in 150 mM NaCl.
-
Figure 2—source data 1
Raw Gel Blots.
- https://cdn.elifesciences.org/articles/96028/elife-96028-fig2-data1-v1.zip
-
Figure 2—source data 2
Labeled Gel Blots.
- https://cdn.elifesciences.org/articles/96028/elife-96028-fig2-data2-v1.zip
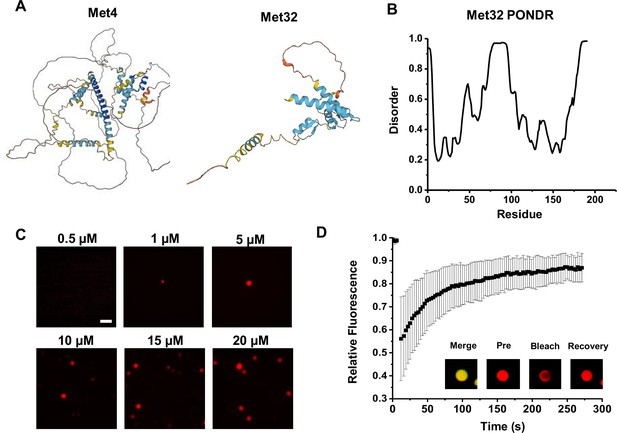
Met4 and Met32 structure and droplet formation.
(A) Predicted protein structures of Met4 (Alphafold: AF-A0A1L4AA63-F1) and Met32 (AF-A0A0L8VTL7-F1). (B) PONDR protein disorder plot of Met32 (same analysis for Met4 is shown in Figure 7). (C) Met32-mCherry condensate formation at various concentrations. Purified Met32-mCherry proteins were titrated in 20 mM HEPES pH 7.5, 150 mM NaCl. Scale bar represents 10 µm. (D) Fluorescence recovery after photobleaching (FRAP) curve for Met4/Met32 co-localized droplets. 5 µM Met4-MBP and 20 µM Met32-mCherry were mixed. Intensity data collected every 3 s for 270 s.
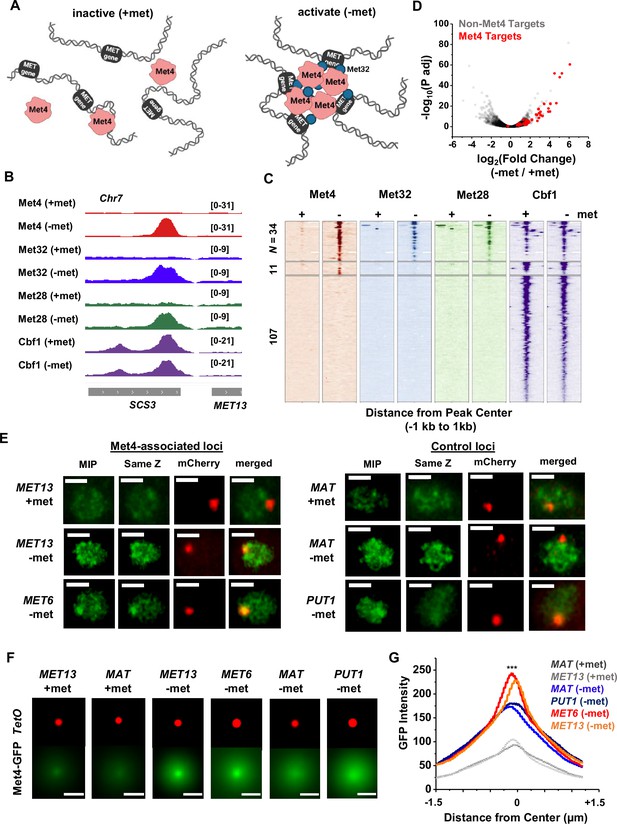
Met4-activated genes co-localize with Met4 puncta.
(A) Model of Met4 condensate in the chromatin context. Upon met depletion, Met transcription factors (TFs) may form condensates that interact with multiple target genes, leading to the 3D clustering of these co-regulated genes. (B) Examples of chromatin immunoprecipitation with sequencing (ChIP-seq) signals of Met TFs (Met4, Met32, Met28, and Cbf1) in ±met conditions. The Met TF co-binding peak shown here is near the MET13 gene. Two biological replicates were performed for each factor. (C) Heatmaps of Met4, Met32, Met28, and Cbf1 ChIP-seq peaks in ±met conditions. The peaks were clustered into ones enriched with all four Met TFs (N = 34), enriched with Met4 and Cbf1 (N = 11), and enriched with Cbf1 only (N = 107). (D) Volcano plot of RNA-seq data comparing mRNA levels in ±met conditions. Most genes near Met TFs co-binding peaks are strongly induced by met depletion (red dots). (E) Single nuclei images of yeast cells expressing Met4-GFP and TetR-mCherry with a TetO array integrated near MET13/MET6 (Met4 targets), or MAT/PUT1 (non-targeted control). Images were taken with 14 z-stacks with step size 0.4 μm. Max intensity projection (MIP) and ‘same Z’ show Met4-GFP images with either maximum intensity projection among all z stacks, or with a single stack at the same z plane as ‘mCherry’, where the mCherry labeled TetO array shows the highest intensity. ‘merged’ is the merged image of the ‘same Z’ and the ‘mCherry’. Scale bars represent 1 µm (same as in F). All strains were imaged three times. (F) MIP of mCherry and GFP intensities. Images of each cell are aligned and centered with the mCherry dot, which represents the TetO array, and the corresponding GFP MIPs for all cells are averaged. Number of cells analyzed: MET13 +/−met (447/413), MET6 (381), MAT +met/−met (290/424), PUT1 (524). (G) The GFP intensity profiles of the averaged MIP images shown in F. A line was drawn across the dot center and the GFP intensity was calculated along the line. The GFP intensity near the dot center is significantly higher for MET13/MET6 loci in −met condition than PUT1 and MAT. Significance was calculated using two-tailed Student’s t-test (***p < 0.001).
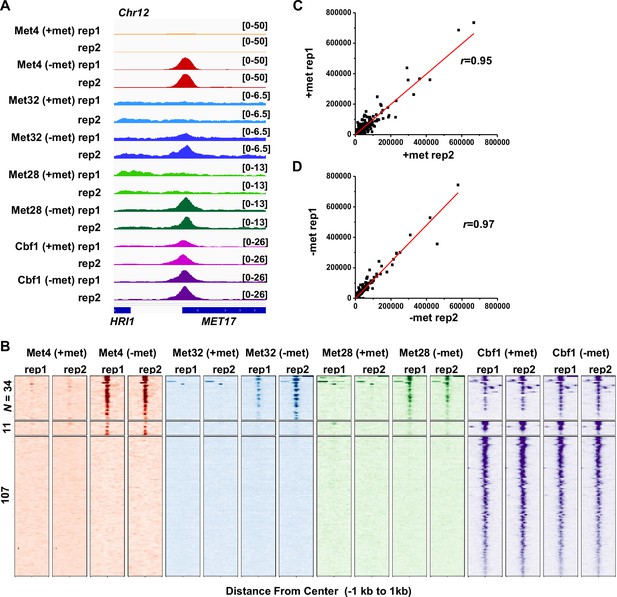
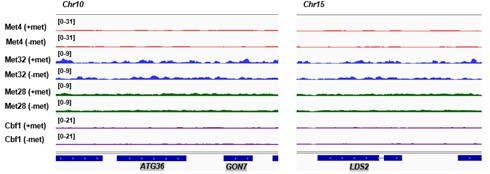
Met TF ChIP-seq and RNA-seq data.
(A) Additional examples of chromatin immunoprecipitation with sequencing (ChIP-seq) data on Met transcription factors (TFs) (Met4, Met32, Met28, and Cbf1). For each factor, two biological replicates in ±met conditions are shown. In this case, the Met TF co-binding peak is in the MET17 promoter. (B) Same as in Figure 3C but showing two replicates of the ChIP-seq data for the four factors in ±met conditions. (C, D) Comparison of read counts from two biological replicates of RNA-seq data in +/−met conditions. Pearson’s correlations (r) between the two biological replicates are shown.
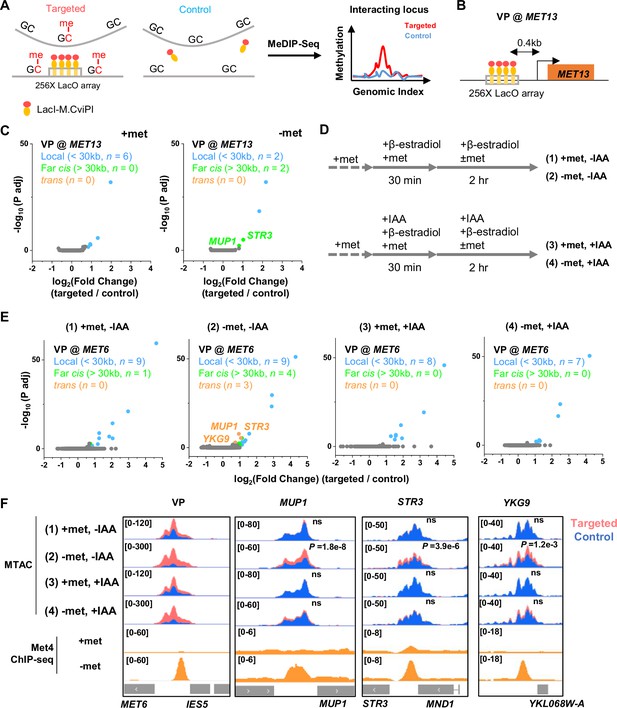
Met4-dependent clustering of MET genes upon induction.
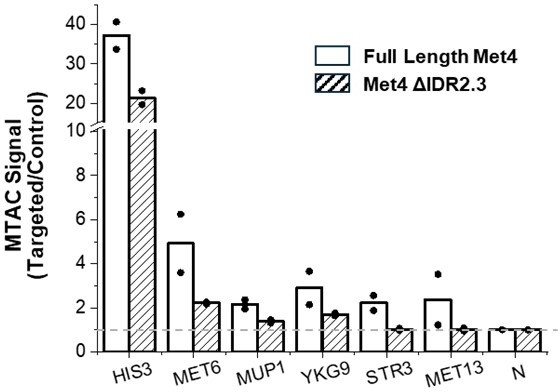
(A) Methyltransferase Targeting-based chromosome Architecture Capture (MTAC) workflow. In a ‘targeted’ MTAC strain, an LacO array is integrated into a genomic locus (viewpoint, VP) and recruits LacI-M.CviPI, an ectopic DNA methyltransferase that methylates the cytosine in a ‘GC’ dinucleotide in proximal DNA. LacI-M.CviPI is also expressed in a control strain with no LacO array insertion (background methylation). Methylation in these two strains is detected by ChIP, and methylation level in nucleosome-depleted regions (NDRs) are compared in targeted vs control strains. Significantly higher methylation in the targeted strain indicates proximity to the VP. (B) VP design of the MET13 locus. (C) Volcano plot of MTAC signals with MET13 as VP in ±met conditions derived from two biological replicates at each condition. Each dot represents an individual NDR, and colored dots are the NDRs that show proximity to the VP (significantly higher methylation in the targeted vs control strains). Local (intra-chromosomal interactions within 30 kb), far-cis (intra-chromosomal interactions over 30 kb), and trans (inter-chromosomal interactions) are shown in blue, green, and orange. Same color scheme is used below. (D) Design of Met4 depletion assay. Met4 is depleted by auxin-degron system in ±met conditions, resulting in four conditions: (1) +met, −IAA, (2) −met, −IAA, (3) +met, +IAA, (4) −met, +IAA. β-Estradiol is added in all conditions to induce the expression of LacI-M.CviPI. (E) Volcano plot of MTAC signals with MET6 as VP for the four conditions in panel D. Note that long-distance interactions are detected in condition (2) but are largely absent in other three conditions. Four biological replicates were performed for each condition. (F) MTAC and Met4 chromatin immunoprecipitation with sequencing (ChIP-seq) data at the MET6 locus (VP) and MUP1, STR3 and YKG9 as interacting regions of the VP. MTAC signal is shown in the four conditions in panel D. The ChIP enrichment of Met4 is shown in ±met conditions. p, False discovery rate (FDR)-adjusted p value, Wald test by DESeq2. ns, non-significant.
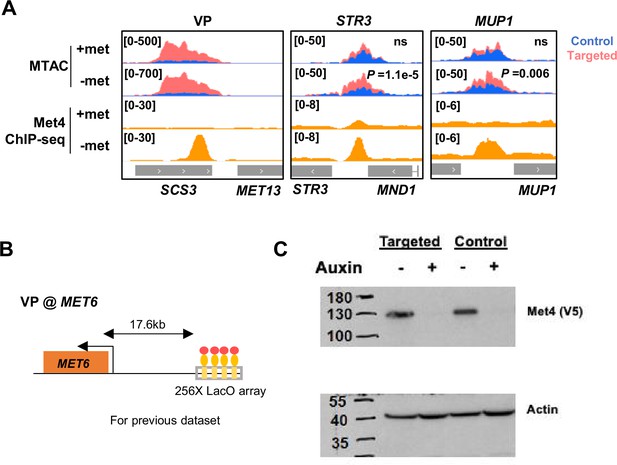
MTAC at MET13 and MET6 loci, and Met4 degradation.
(A) Methyltransferase Targeting-based chromosome Architecture Capture (MTAC) and Met4 chromatin immunoprecipitation with sequencing (ChIP-seq) data at the MET13 locus (VP), as well as at STR3 and MUP1 as interacting regions of the VP. MTAC signal and ChIP enrichment are both shown in ±met conditions. p, FDR-adjusted p value, Wald test by DESeq2. ns, non-significant. (B) VP design near the MET6 locus used for Figure 4D–F. (C) Western blot of V5-AID-tagged Met4 ±Auxin (IAA) in MTAC targeted and control strains. Actin is probed as input loading control.
-
Figure 4—figure supplement 1—source data 1
Auxin induced degradation of Met4.
- https://cdn.elifesciences.org/articles/96028/elife-96028-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Raw western blot of auxin degraded Met4.
- https://cdn.elifesciences.org/articles/96028/elife-96028-fig4-figsupp1-data2-v1.zip
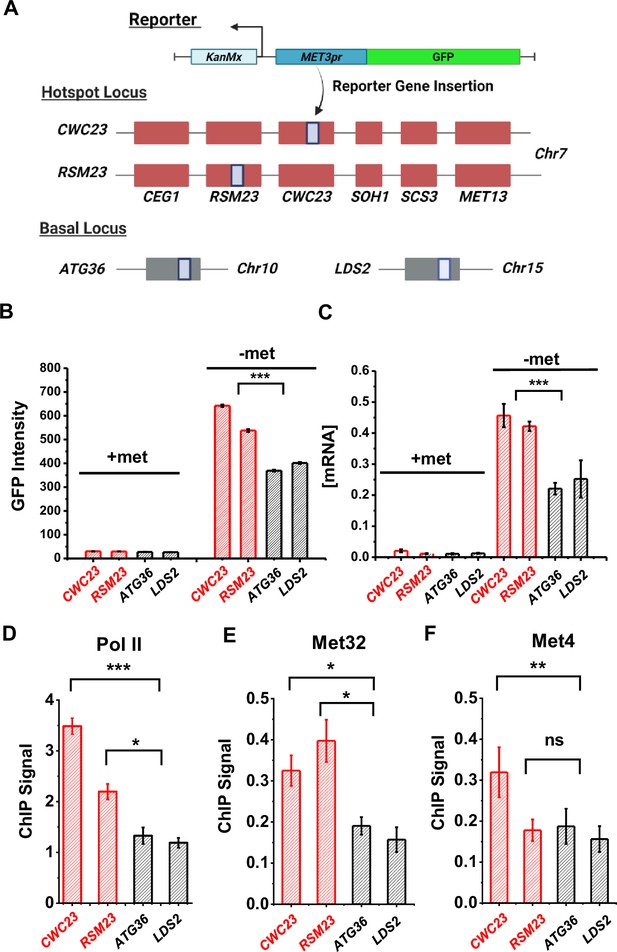
Characterization of a MET ‘transcriptional hotspot’ in haploid yeast.
(A) Schematic of strains constructed with S.kud MET3pr-GFP reporter inserted near the MET13 transcriptional hotspot (CWC23, RSM23) and ‘basal’ loci (ATG36, LDS2). (B, C) Mean cellular GFP fluorescent intensities and GFP mRNA measured by qRT-PCR in the four strains above in ±met conditions. Error bars represent standard error among three biological replicates, and the asterisks represent *<0.05, **<0.01, ***<0.001 (same for D–F). (D) ChIP-qPCR of Rpb1 over the GFP ORF in the four strains above in −met. (E, F) ChIP-qPCR of Met32 and Met4 over the S.kud MET3pr in the four strains above in −met.
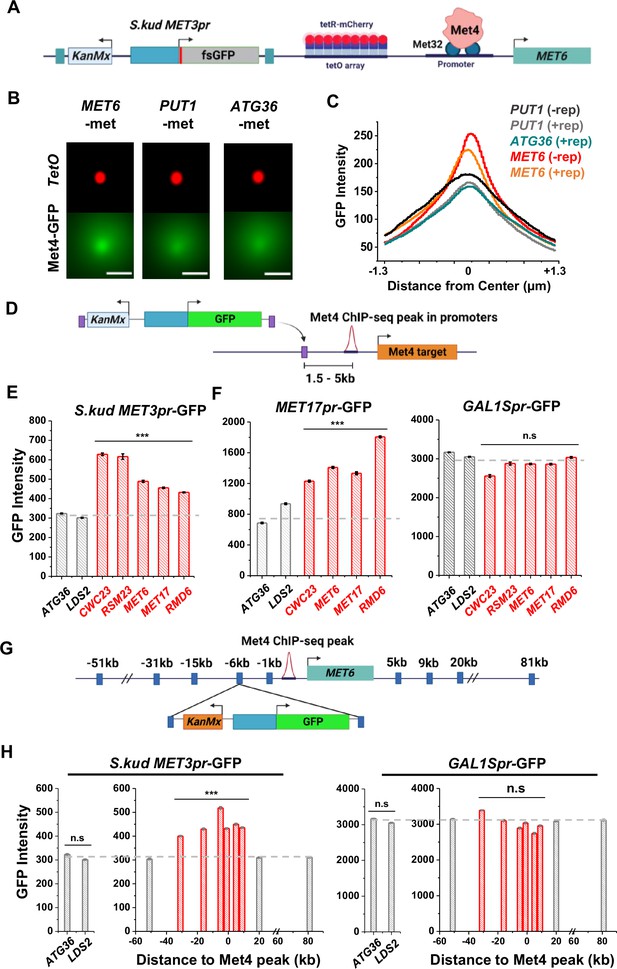
Reporter activities near Met4-binding sites are enhanced over a ~40-kb range.
(A) Schematics of measuring the co-localization of the GFP reporter with Met4 puncta. S.kud MET3pr-fsGFP (frameshifted GFP) reporter gene and a tetO array (196x) are inserted side by side into the genome, in this case near the MET6 gene. (B) Averaged Max intensity projections (MIPs) of mCherry and GFP intensities near MET6 (Met4 target) and PUT1/ATG36 (not Met4 targets) in the presence of nearby reporter. These images are generated using the same method as in Figure 3F. Scale bars represent 1 µm. (C) The GFP intensity profile across the dot center in panel B. For MET6 and PUT1, the same type of data without the GFP reporter (−rep) are also included. Number of cells analyzed (for panels B and C): PUT1 +/−rep (318/524), ATG36 +rep (386), and MET6 +/−rep (330/381). (D) Schematic of GFP reporter insertion near three additional Met4-bound loci, MET17, RMD6, and MET6. The distance and orientation of the insertion are labeled in the diagram. (E) Mean GFP fluorescent intensities when S.kud MET3pr-GFP are inserted near indicated genes. The genes labeled in red have adjacent Met4-bound sites, while the ones labeled in gray do not. Error bars represent standard error among cells (collected in three independent experiments), and the asterisks *** represents P<0.001 (same for F, H). (F) Same as in panel E except with MET17pr-GFP and GAL1Spr-GFP reporter. Strains with MET17pr-GFP were grown in −met, and the ones with GAL1Spr-GFP were pre-grown in raffinose and induced by galactose for 6 hr. (G) Schematic of the MET3pr-GFP reporter inserted at various distances from the Met4-binding site near the MET6 gene. The same orientation was used for all the loci as indicated. (H) Mean GFP fluorescent intensities with MET3pr-GFP and GAL1Spr-GFP reporters inserted into locations indicated in panel G, in comparison to the same reporter inserted into two control loci far from Met4-binding sites.
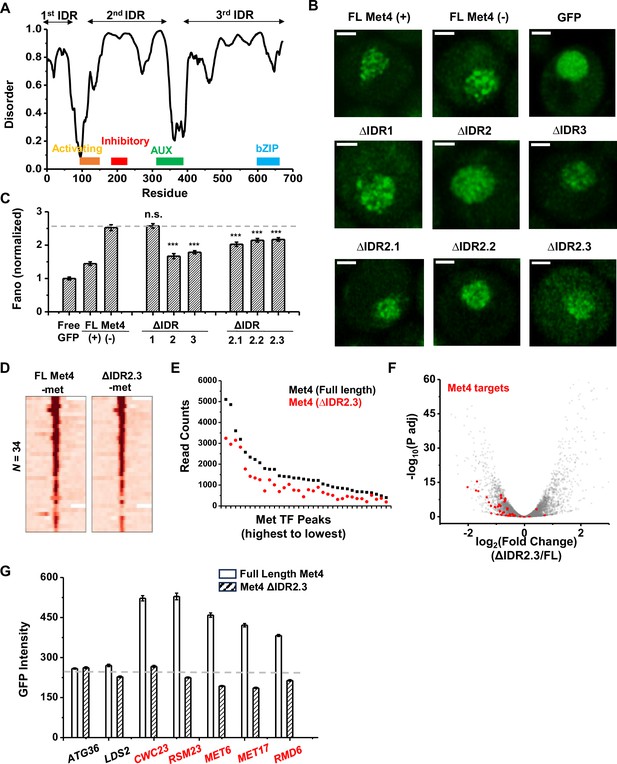
The deletion of a disordered region in Met4 reduces puncta formation and reporter expression at transcriptional hotspots.
(A) PONDR disorder plot of Met4 with previously annotated functional domains, including activation domain (aa95–144) that interacts with Mediator, inhibitory domain (aa188–235), auxiliary domain (aa312–375) that interacts with Met31/32, and bZIP domain (aa595–660) that interacts with Cbf1. (B) Single nuclei images of cells expressing full-length Met4 (±met) or Met4 with various truncations (−met) fused with GFP. Images were collected four times. (C) The Fano numbers of nuclear pixel intensities for different versions of GFPs in panel B. Number of cells analyzed: Met4GFP +/−met (65/147), Free GFP (67), ΔIDR1/2/3 (172/95/172), ΔIDR2.1/2.2/2.3 (134/177/161). Error bar represents standard error among all cells. The asterisks *** represents P<0.001 (D) Heatmaps of full-length Met4 and Met4 ΔIDR2.3 chromatin immunoprecipitation with sequencing (ChIP-seq) in −met over previously identified Met transcription factor (TF)-binding sites. Two biological replicates were collected. (E) Alignment counts underneath the Met TF ChIP-seq peaks over the same sites for TAP-tagged full-length Met4 and Met4 ΔIDR2.3 (sorted from high to low). (F) Volcano plot from two biological replicates of RNA-seq data from cells containing full-length Met4 or Met4 Δ IDR2.3 in −met. Genes associated with Met TFs (Figure 3C) are indicated in red. (G) Mean GFP intensities from cells expressing full-length Met4 and Met4 ΔIDR2.3 with MET3pr-GFP reporter inserted into loci near (red) or far away from (black) Met4-target genes. The dashed lines represent basal reporter expression. Error bar represents standard error among cells (collected in three independent experiments).
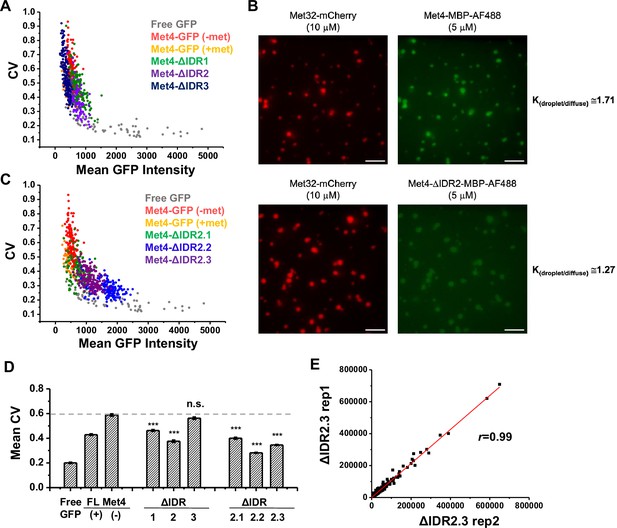
Disordered regions of Met4 affect its puncta formation.
(A) Coefficient of variance (CV) of pixel intensities vs average intensity in cells expressing free GFP or GFP fused with full-length Met4 (in ±met), ΔIDR1 (aa1–69), ΔIDR2 (aa117–359), and ΔIDR3 (aa397–651). Individual dots represent single nuclei. IDR strains were grown in SCD-met. (B) Partitioning of full-length Met4 and Met4-ΔIDR2 into the Met32 condensates. The partition coefficient (K), measuring the ratios of the green fluorescent intensity inside vs outside the Met32 droplet are shown on the right. (C) Same as in A except with Met4 ΔIDR2.1 (aa117–135), ΔIDR2.2 (aa136–270), and ΔIDR2.3 (aa271–359). IDR strains were grown in SCD-met. (D) The mean CV values of nuclear pixel intensities of different GFP fusions in panels A and B. p values were calculated against the Free GFP. The asterisks represent *** = P<0.001(E) Comparison of read counts from two biological replicates of RNA-seq data of the ΔIDR2.3 strain in −met. Pearson’s correlations (r) between the two biological replicates are shown.
Tables
| FPKM counts | Rep1 (+met) | Rep2 (+met) | Rep1 (-met) | Rep2 (-met) |
|---|---|---|---|---|
| ATG36 | 6.15 | 13.59 | 6.56 | 7.15 |
| LDS2 | 5.35 | 32.44 | 2.24 | 2.99 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96028/elife-96028-mdarchecklist1-v1.pdf
-
Supplementary file 1
List of yeast strains, plasmids, and primer sequences.
- https://cdn.elifesciences.org/articles/96028/elife-96028-supp1-v1.xlsx
-
Supplementary file 2
Raw GFP signal per cell, including average intensity and standard deviation among pixels.
- https://cdn.elifesciences.org/articles/96028/elife-96028-supp2-v1.xlsx
-
Supplementary file 3
ChIP-seq peak coordinates of TFs in the MET regulon.
- https://cdn.elifesciences.org/articles/96028/elife-96028-supp3-v1.xlsx
-
Supplementary file 4
RNA-seq counts of Met TF associated genes.
- https://cdn.elifesciences.org/articles/96028/elife-96028-supp4-v1.xlsx