Lymphoid origin of intrinsically activated plasmacytoid dendritic cells in mice
Figures
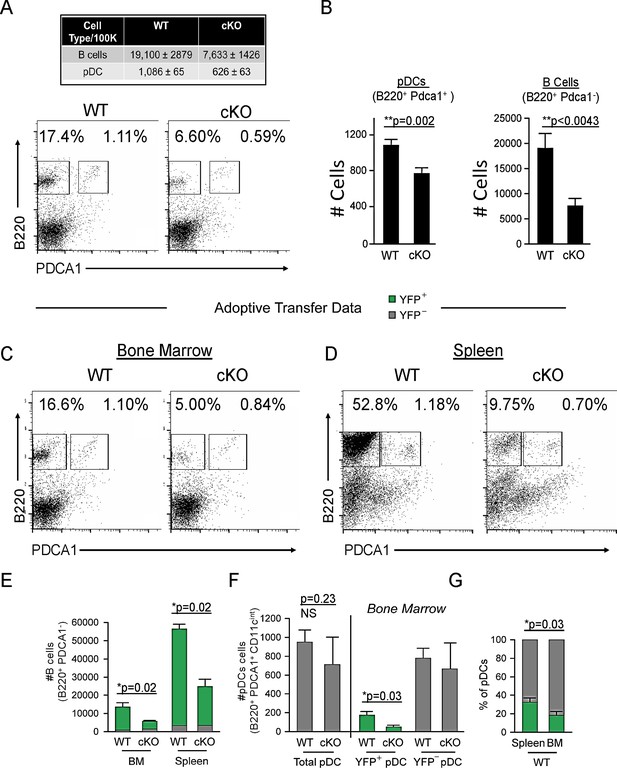
Cd79a-Cre deletion of Bcl11a identifies a common lymphoid progenitor (CLP)-derived subset of plasmacytoid dendritic cells (pDCs).
(A) Representative FACS plots of pDC (gated as B220+ PDCA1+) and B cell (gated as B220+ PDCA1-) percentages in the bone marrow (BM) of Bcl11a F/F Cd79a-Cre mice (cKO) and littermate controls. (B) Quantification of pDC and B cell populations in the BM of Bcl11a F/F Cd79a-Cre mice (cKO) and littermate controls as cells/100,000 cells. (C–D) Flow cytometric analysis of BM and spleens of recipient mice 8 weeks post BM transplantation. BM was transferred from either BCL11A-sufficient reporter control mice (Cd79a-Cre-YFP) or BCL11A-deficient cKO mice (Bcl11a F/F Cd79a-Cre-YFP) into lethally irradiated C57BL/6J recipients. (E) B cell numbers after BM transplantation in BM and spleens of recipient mice. (F) pDC numbers after BM transplantation in BM and spleens of recipient mice. (G) Comparison of YFP+ pDC percentages in the spleen and BM of recipient mice post BM transplantation. Mann-Whitney t-tests were used for all statistical comparisons. Error bars=mean±s.d. The results are representative of two experiments each containing 3–4 mice per group.
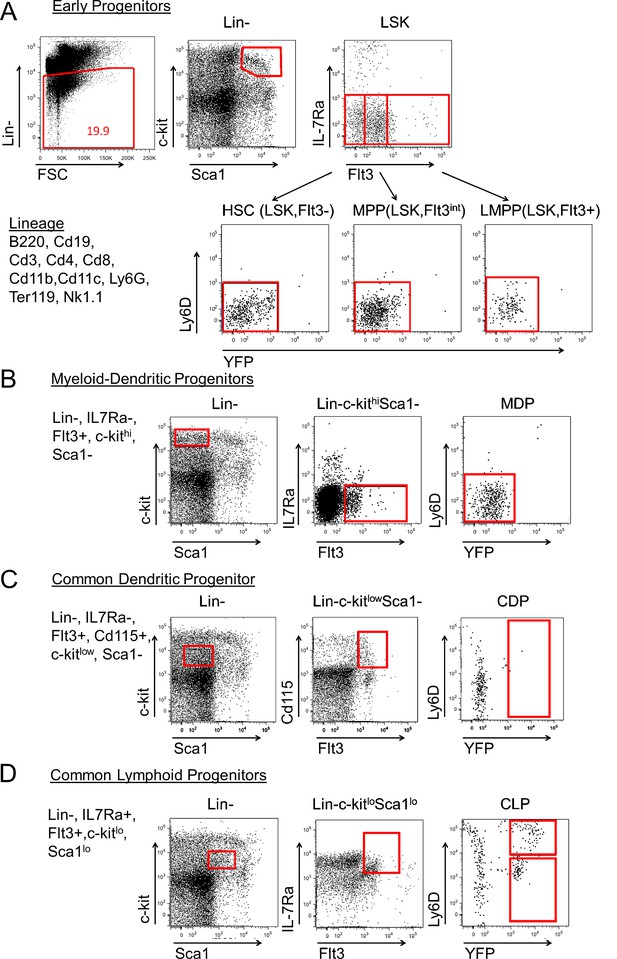
Progenitor population analysis for Cd79a-cre driven YFP expression.
We analyzed YFP expression in bone marrow (BM) progenitors to determine when and where Cd79a-Cre is active (representative plots shown). (A) Lineage negative cells were analyzed to examine LSK hematopoietic progenitor (Lin-, Sca-1+, c-Kit+), MPP (LSK, Flt3int), and LMPP (LSK, Flt3hi) populations. Virtually all cells were YFP-. (B) Myeloid dendritic progenitors (MDPs; Lin-, IL7Rα-, Flt3+, c-kithi, Sca1-) and (C) common dendritic cell progenitors (CDPs; Lin-, IL7Ra-, Flt3+, c-kitlo, Sca1-, CD115+) were also negative for YFP expression, while (D) common lymphoid progenitors (CLPs; Lin-, IL7Ra+, Flt3+, c-kitlo, Sca1lo) contained YFP+ cells. The results are representative of three independent experiments each containing at least 3 mice per group.
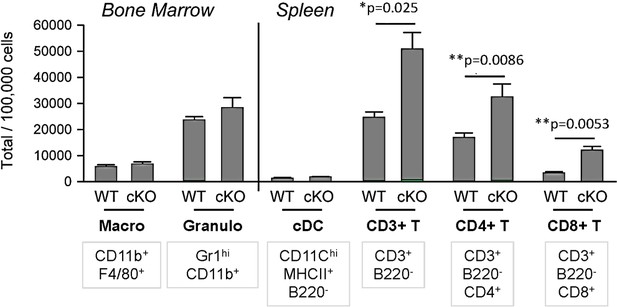
Cell distributions in the spleen of adoptively transferred recipient mice.
Both Bcl11a F/F Cd79a-Cre+ and Cd79a-Cre-YFP adoptively transferred recipient mice reconstituted other splenic cell types in normal numbers, including bone marrow (BM) macrophages (CD11b+ F4/80+), granulocytes (Gr-1+ CD11b+), and splenic conventional dendritic cells (cDCs) (CD11c+ CD11b+ B220-). Total T cells (CD3+ B220-) as well as CD4+ and CD8+ subsets were significantly increased in number in proportion to B/pDC cell loss. Mann-Whitney t-tests were used for all statistical comparisons. The results are representative of two independent experiments each containing 3–4 mice per experimental group. Error bars = mean ± s.d.
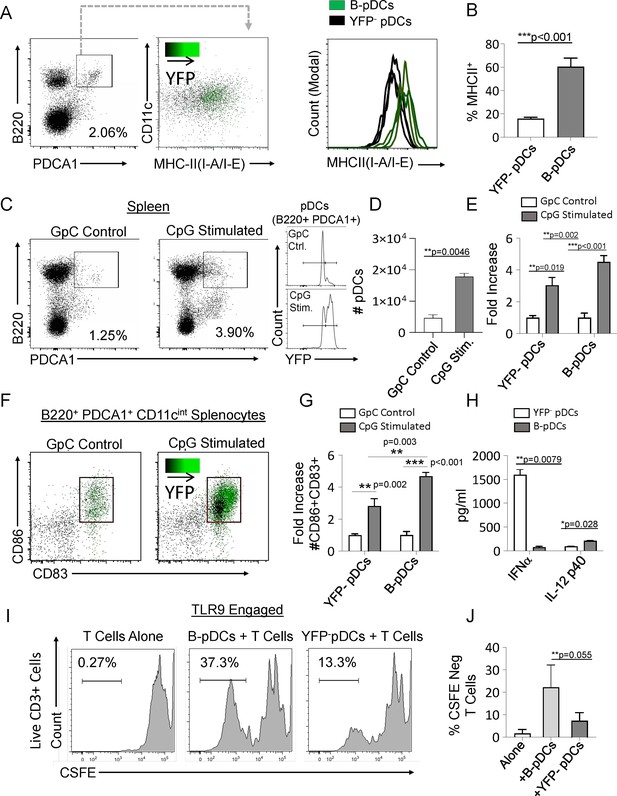
Common lymphoid progenitor (CLP)-derived B-plasmacytoid dendritic cell (B-pDC) are licensed for T cell activation.
(A–B) Comparison of MHCII MFI in YFP + and YFP- pDCs from Cd79a-Cre-YFP reporter mice via flow cytometric analysis. (C–D) Cd79a-Cre-YFP mice were injected with 50 µg/mL (100 µL) of CpG:ODN or control GpC:ODN and analyzed via flow cytometry for splenic pDC numbers/100,000 cells. (E) Flow cytometry quantification of YFP- pDC and B-pDCs (YFP+) fold change in cell numbers upon CpG:ODN in vivo challenge. (F–G) Fold change in CD86+CD89+ cell numbers for each pDC population in mice restimulated with CpG:ODN. Difference in CD86+CD89+ cell numbers between stimulated B-pDCs and YFP- pDCs was also significant. (H) In vitro Toll-like receptor 9 (TLR9) engagement of B-pDC or pDC for ELISA against IFN-α orIL-12p40. (I) T cells were magnetically isolated from wildtype C57BL/6J mouse splenocytes using MACs columns, labeled with CSFE, and then cultured (2.5×104/well) alone or with CpG:ODN activated B-pDCs or pDCs (5×103/well) for 6 days. (J) The percentage of CFSE-negative CD3+ T cells in co-cultures were significantly higher in B-pDC compared to pDCs. Mann-Whitney t-tests were used for all statistical comparisons. Error bars = mean ± s.d. The results are representative of three independent experiments each containing at least 4 mice per group.
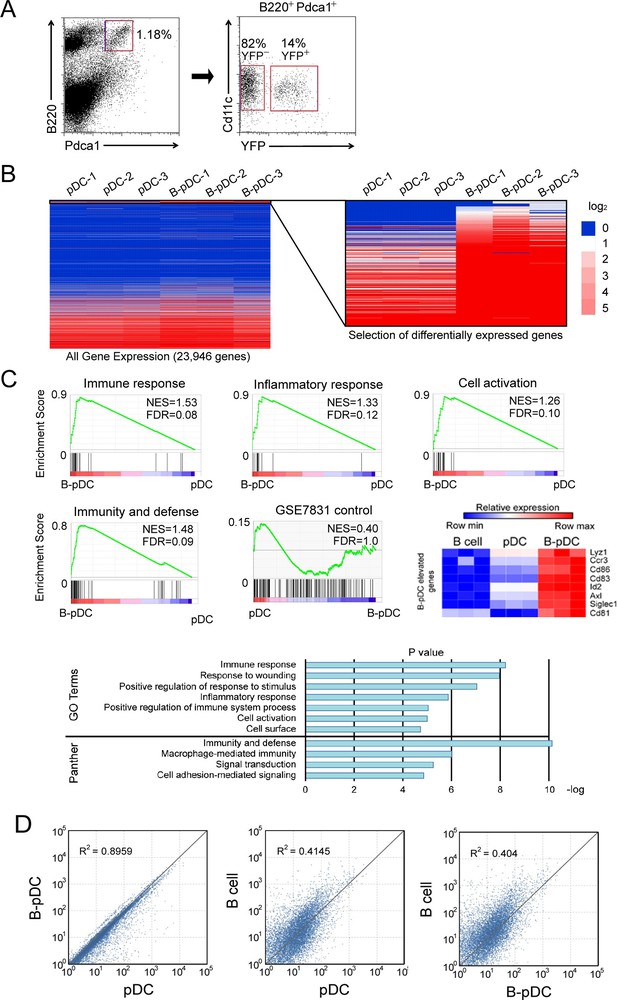
Transcriptional analysis identifies two populations of plasmacytoid dendritic cell (pDC) in mice: myeloid-derived classical pDC and common lymphoid progenitor (CLP)-derived B-pDC.
(A) Bone marrow pDC (B220+ PDCA1+ CD11cint CD11b-) were sorted based on expression of YFP. Four mice were pooled for each isolated RNA sample, for a total of three pDC and three B-pDC groups from 12 mice. (B) RNA-seq was performed for gene expression analysis of pDC vs. B-pDC and 220/23,946 genes (~1%, left) were significantly differentially expressed (q-value<0.05, right heatmap, Log2 expression difference displayed). (C) Gene set enrichment analysis (GSEA). Normalized enrichment score (NES) and false discovery rate q-values (FDR); FDR≤.25 is considered significant (Mootha et al., 2003). GO term or Panther-derived pathways identified by DAVID analysis of ~220 differentially expressed genes (DEGs) between the pDC and B-pDC subsets (Thomas et al., 2003; Liu et al., 2014). Among the top B-pDC DEGs were Lyz1, Ccr3, Cd86, Cd83, Id2, Axl, Siglec1, and Cd81. (D) Scatter plot comparisons of all genes with reads per kilobase of transcript, per million mapped reads (RPKM) >1. Correlations of pDC vs. B-pDC (R2 value = 0.8959), B cell vs. pDC (R2 value = 0.4145), and B cell vs. B-pDC (R2 value = 0.404) are indicated.
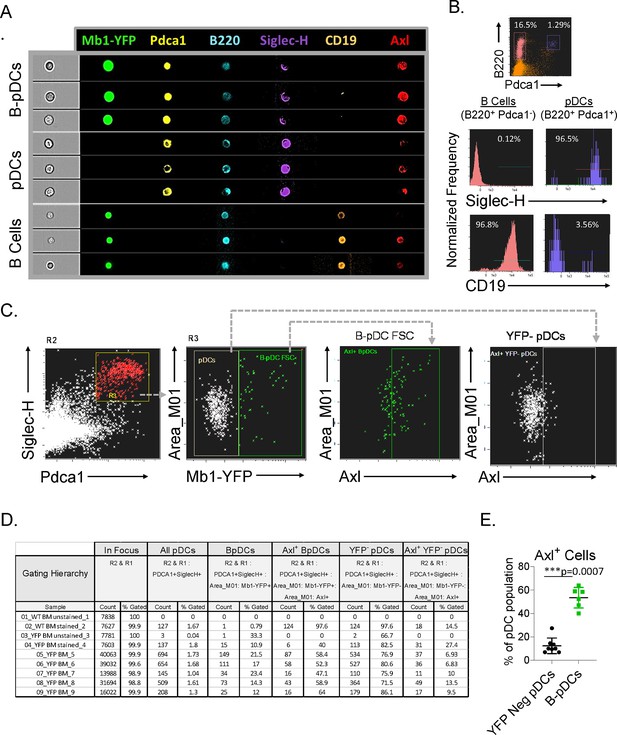
Imaging flow cytometry as a tool to quantify AXL expression in plasmacytoid dendritic cell (pDC) populations.
(A–B) Representative imaging flow cytometry of manuscript gating scheme used to define B cells and pDCs and histograms for SIGLEC-H, AXL, and CD19 in B cells and pDCs. (C) Representative imaging flow cytometry plots of AXL+ pDCs (here alternatively gated as SIGLEC-H+ PDCA1+) in Cd79a-Cre-YFP reporter mice bone marrow cells (n=5). (D) Complete statistics file generated with IDEAS for ImageStream X analysis including non-stained wildtype controls. (E) Comparison of AXL expression in YFP- and YFP+ (B-pDCs). The results are representative of two independent experiments each containing at least 3 mice per group. Mann-Whitney tests were used as statistical method.
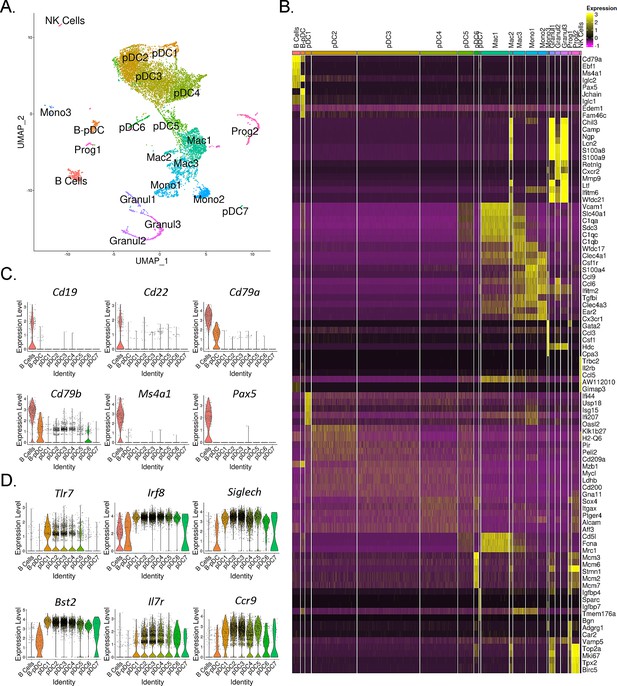
Single-cell RNA-seq analysis of mouse PDCA1+ bone marrow cells.
(A) UMAP generated by Seurat clustering analysis. Cluster identities were assigned using the top 5 consensus CIPR (Cluster Identity Predictor) identity scores. (B) Heatmap of the top 5 differentially expressed genes (DEGs) from each cluster. After Seurat clustering, cell reads were subsetted to include only cells classified as B cells, plasmacytoid dendritic cells (pDCs), and B-pDCs. DEGs were generated for the new data subset and markers associated with (C) mature B cells and (D) classical pDCs were plotted as violin plots.
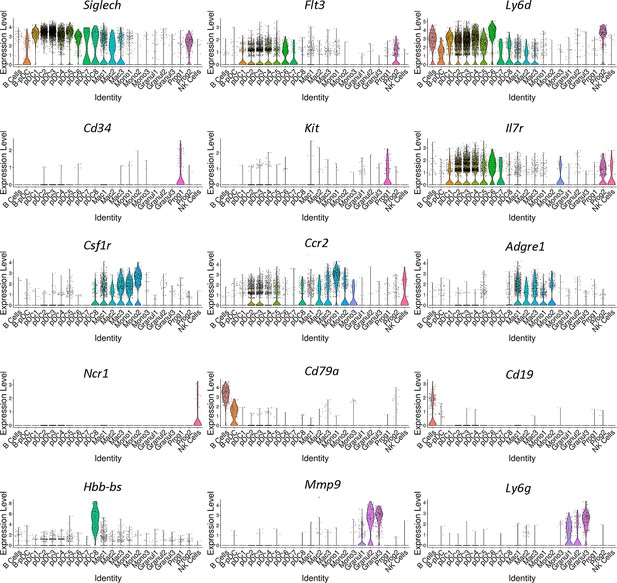
Confirmation of unbiasedly identified cell cluster identities.
Seurat generated violin plots of genes associated with distinct cell populations including mature B cells, plasmacytoid dendritic cells (pDCs), granulocytes, pre-pDCs, NK cells, monocytes, macrophages, and myeloid progenitor cells. Plot identities were initially determined by running the package CIPR (Cluster Identity Predictor).
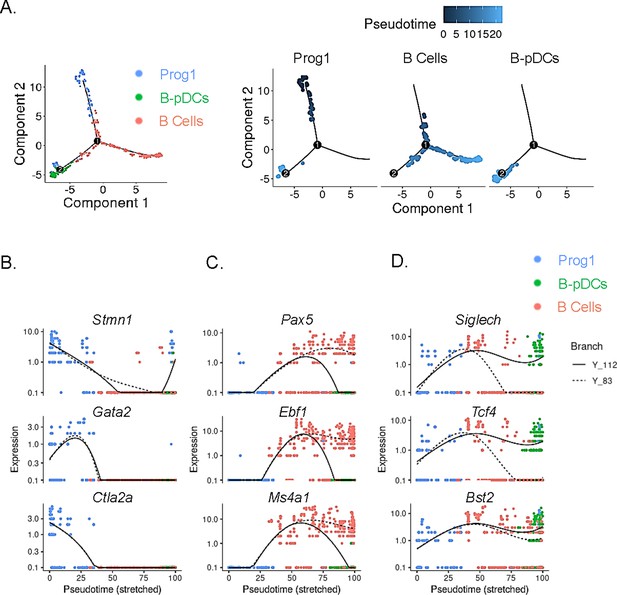
Single-cell trajectory analysis of Prog1, B cells, and B-plasmacytoid dendritic cell (B-pDC) clusters.
(A) Single cells belonging to clusters Prog1, B cells, and B-pDCs were ordered and plotted as a function of pseudotime based on uniquely expressed markers using the unsupervised Monocle dpFeature. Cluster Prog1 was classified as the root of the trajectory given its high expression of pre-pro B cell and stem cell associated markers. Differentially expressed genes (DEGs) of interest were plotted as a function of pseudotime in Prog1 (B), B cells (C), or pDCs (D) using Monocle. Branch Y_112 represents the expression kinetic trend of the B-pDC cluster (green), while branch Y_83 represents the expression kinetic trend in expression of B cells according to branched expression analysis modeling, or BEAM.
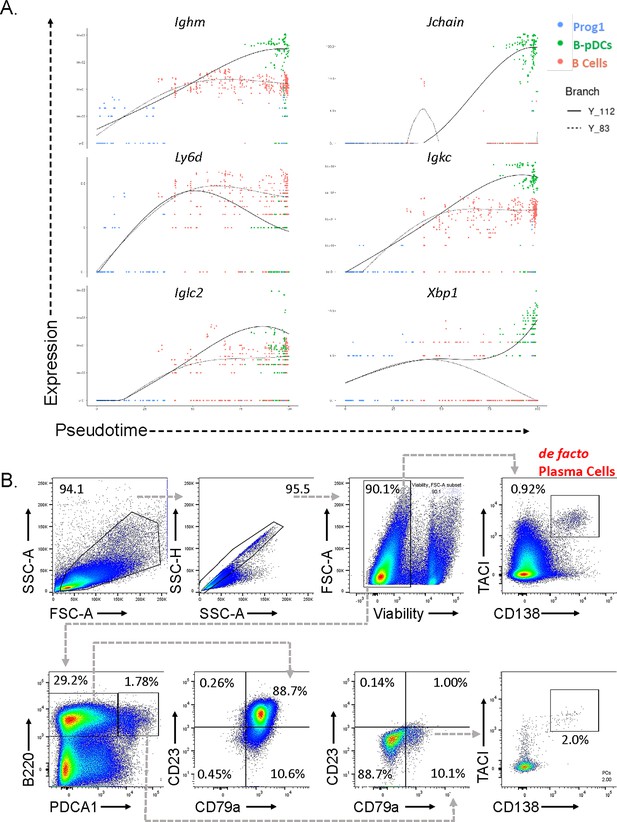
Expression of early B cell receptor genes in B-plasmacytoid dendritic cells (B-pDCs).
(A) Monocle was used to plot select early B cell genes as function of branched pseudotime for Prog1, B cells, and B-pDC populations. (B) Plasma cell contamination in Cd79a+ pDCs (B220+ PDCA1+) was ruled out by flow cytometric analysis of TACI and CD138 in bone marrow (BM) of 6 C57BL/6J mice.
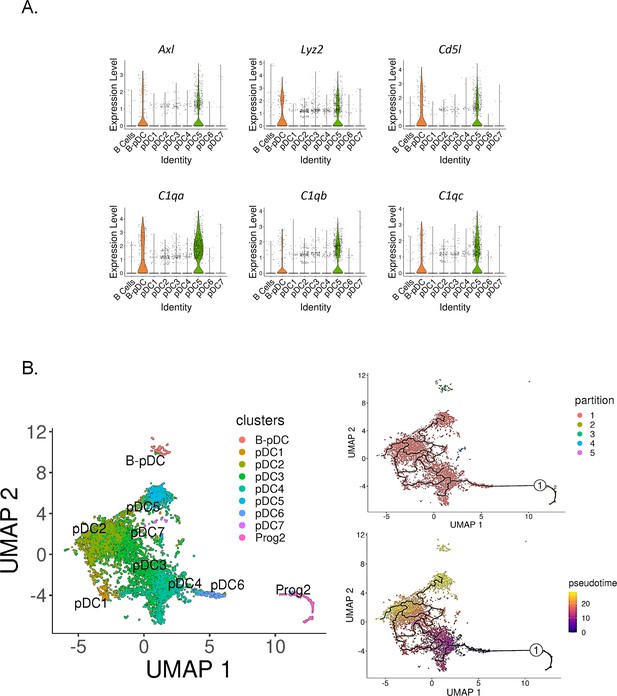
Identification of Axl+ noncanonical plasmacytoid dendritic cells (pDCs) in mice.
(A) Violin plots depicting relative expression of conserved ‘non canonical AXL+ DCs’ associated markers in mouse pDC clusters. (B) Bone marrow (BM) scRNA-seq data was subsetted to include only pDCs and Prog2 subsets. We used Monocle3 for partition-based learning of cell developmental trajectories. Louvain partitions were generated from single-cell reads and their developmental trajectory was inferred using SimplePPT (top right quadrant). Pseudotime across clusters is shown in the lower bottom quadrant.
Additional files
-
Supplementary file 1
Top CIPR (Cluster Identity Predictor) assigned identity scores for Seurat cluster differentially expressed genes (DEGs).
CIPR IDs were generated using the top 1000 DEGs from each cluster. The top 5 consensus CIPR ID generated was used to rename Seurat clusters.
- https://cdn.elifesciences.org/articles/96394/elife-96394-supp1-v1.xlsx
-
Supplementary file 2
Differentially expressed gene (DEG) comparison between progenitor clusters.
The top 50 differentially expressed features of Prog1 in relation to Prog2 were calculated using Seurat using non-parametric Wilcoxon rank sum test. In blue are upregulated genes and in red are downregulated genes.
- https://cdn.elifesciences.org/articles/96394/elife-96394-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96394/elife-96394-mdarchecklist1-v1.docx





