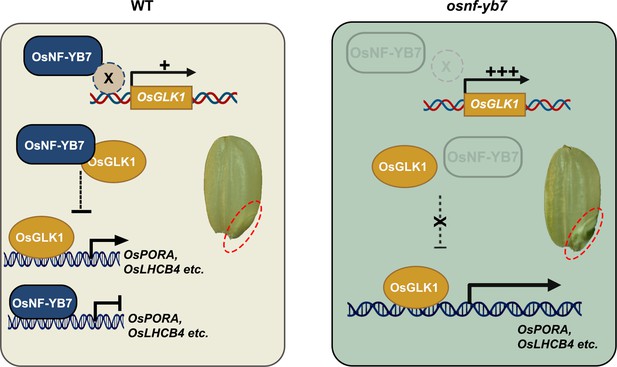
OsNF-YB7 inactivates OsGLK1 to inhibit chlorophyll biosynthesis in rice embryo
Figures
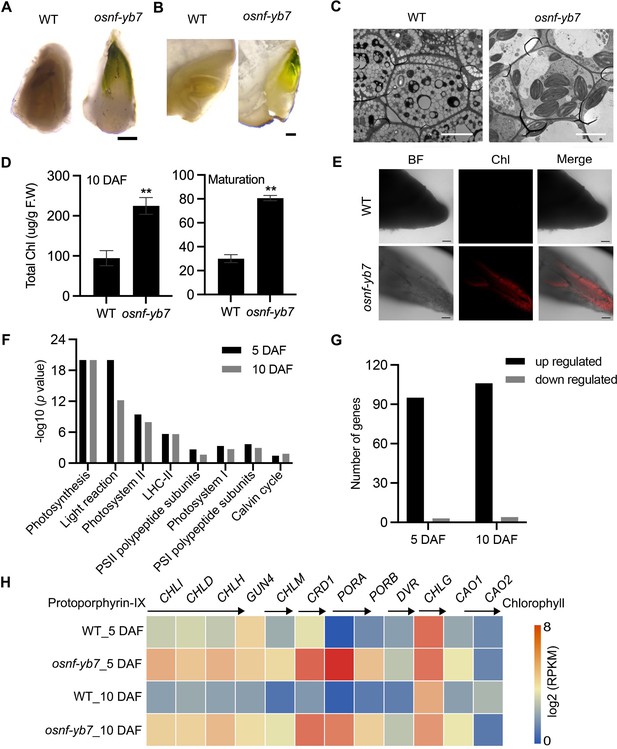
OsNF-YB7 negatively regulates Chlorophyll (Chl) biosynthesis in embryo.
(A, B) Morphologies of wild-type (WT) and osnf-yb7 detached embryos at 10 days after fertilization (DAF) (A) and longitudinally dissected embryos at maturation (B). Scale bars = 200 μm. (C) Transmission electron microscopy images of embryos from WT and osnf-yb7 at 10 DAF. Scale bars = 5 μm. (D) Chl levels in WT and osnf-yb7 embryos at 10 DAF and maturation. Data are means ± SD (n=3). **, p<0.01; Student’s t-test was used for statistical analysis. (E) Chl autofluorescence of WT and mutant embryos at 5 DAF. BF, bright field; Chl, Chl autofluorescence. Scale bars = 100 μm. (F) Photosynthesis-related pathways enriched among the differentially expressed genes (DEGs) identified from the 5- and 10-DAF-old osnf-yb7 embryos compared to WT, respectively. (G) Most of the photosynthesis-related genes were upregulated in the mutant embryos. (H) A heat map shows the expression of the Chl biosynthesis-related genes in the WT and osnf-yb7 embryos at 5 and 10 DAF. Reads per kilobase per million mapped reads (RPKM) is used to indicate the expression level.
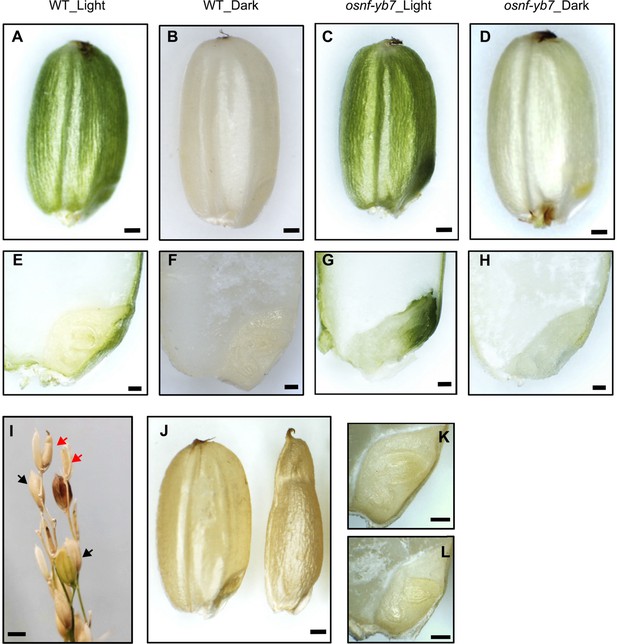
Light is required but not sufficient for Chl biosynthesis in the chloroembryo of osnf-yb7.
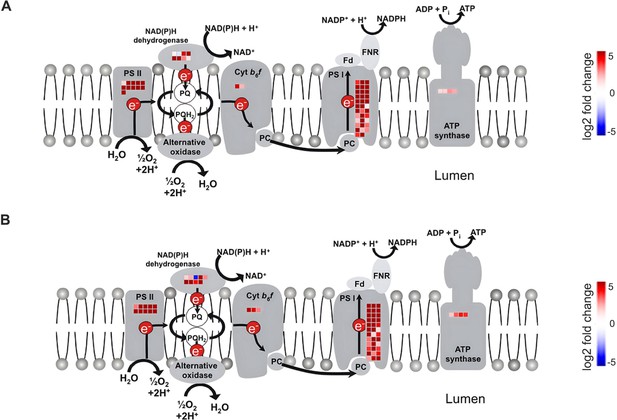
Mutation of OsNF-YB7 actives the expression of photosynthesis-related genes.
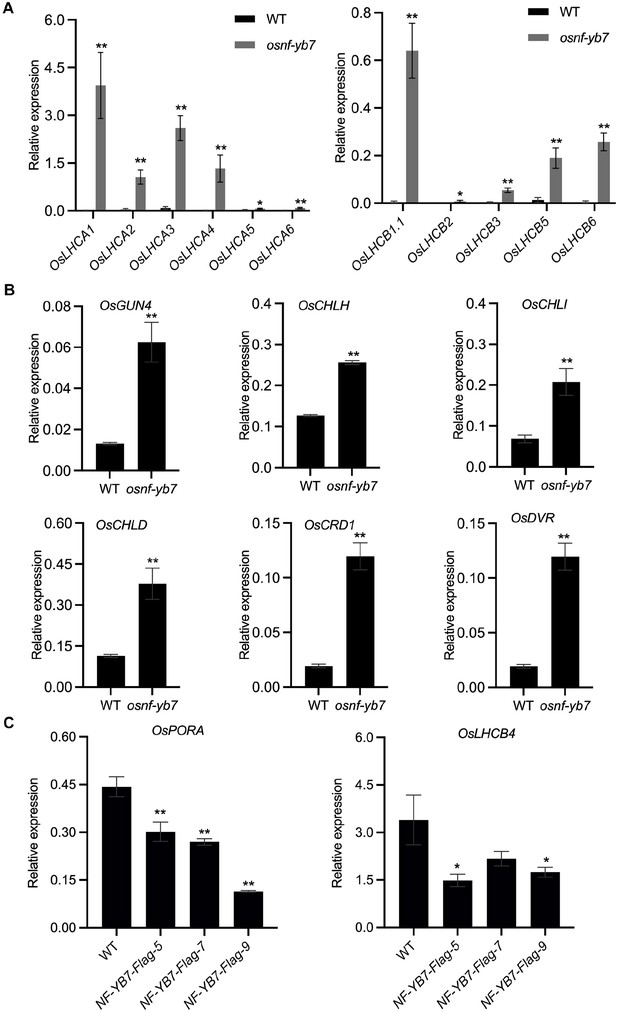
OsNF-YB7 negatively regulates the expression of light harvest and Chl biosynthesis-associated genes.
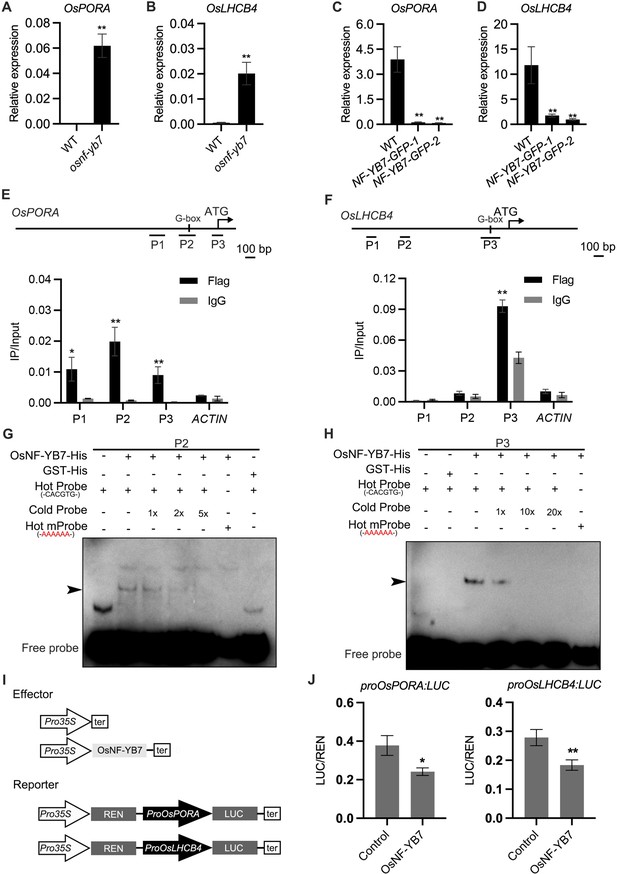
OsNF-YB7 binds to the promoters of OsPORA and OsLHCB4 to regulate their expression.
(A, B) Quantitative real-time PCR (RT-qPCR) analysis of the transcription levels of OsPORA (A) and OsLHCB4 (B) in the embryos of WT and osnf-yb7 at 10 DAF. Data are means ± SD (n=3). **, p<0.01; Student’s t-test was used for statistical analysis. (C, D) Expression of OsPORA (C) and OsLHCB4 (D) in the leaves of WT and OsNF-YB7-overexpressing transgenic plants (NF-YB7-GFP). Data are means ± SD (n=3). **, p<0.01; Student’s t-test was used for statistical analysis. (E, F) Chromatin immunoprecipitation assay coupled with quantitative PCR (ChIP-qPCR) analyses showing the enrichment of OsNF-YB7 at the OsPORA (E) and OsLHCB4 (F) promoters in 14-day-old OsNF-YB7-Flag seedlings. Precipitated DNA was quantified by qPCR and DNA enrichment is displayed as a percentage of input DNA. Data are means ± SD (n=3). *, p<0.05; **, p<0.01; Student’s t-test was used for statistical analysis. ACTIN was used as a nonspecific target gene. Diagrams in the upper panel showing the promoter structures of OsPORA and OsLHCB4, and the PCR amplicons used for ChIP-qPCR. (G, H) Electrophoretic mobility-shift assays (EMSAs) showing that OsNF-YB7 directly binds to the promoters of OsPORA (G) and OsLHCB4 (H). Hot probes were biotin-labeled. The hot mProbes contain mutant nucleic acid from CACATG to AAAAAA. The arrow heads indicate the shift bands. (I) Schematic diagram displaying the constructs used in the dual luciferase reporter (DLR) assays of J. LUC, firefly luciferase; REN, Renilla luciferase. (J) DLR assays showing that OsNF-YB7 directly represses the promoter activities of OsPORA and OsLHCB4. Data are means ± SD (n=3). *, p<0.05; **, p<0.01; Student’s t-test was used for statistical analysis.
-
Figure 2—source data 1
Uncropped and labelled gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig2-data1-v1.zip
-
Figure 2—source data 2
Raw unedited gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig2-data2-v1.zip
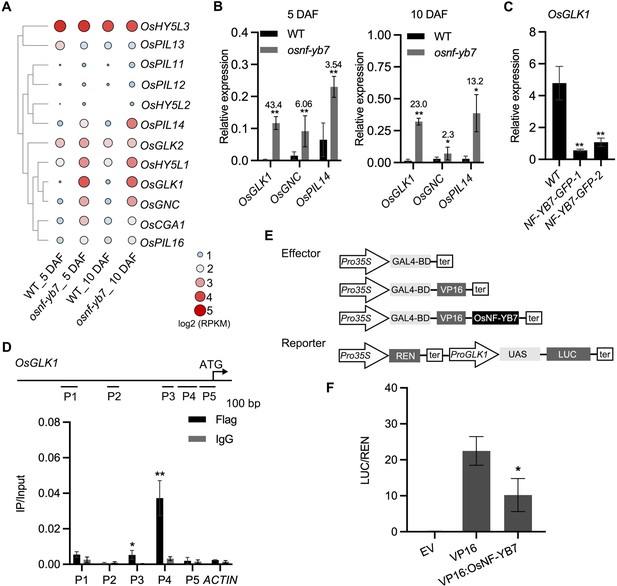
OsNF-YB7 associates with the promoter of OsGLK1 and represses its expression.
(A) A heat map showing the expression of transcription factors associated with Chl biosynthesis and chloroplast development in the 5- and 10-DAF-old embryos of WT and osnf-yb7. The colored dots indicate log2(RPKM mean) of the genes in three biological replicates. (B) RT-qPCR analysis of OsGLK1, OsGNC, and OsPIL14 expression levels in the embryos from WT and osnf-yb7 at 5- and 10 DAF. Numbers represent fold changes of expression. Data are means ± SD (n=3). *, p<0.05; **, p<0.01; Student’s t-test was used for statistical analysis. (C) RT-qPCR analysis of OsGLK1 expression levels in leaves from WT and NF-YB7-GFP. Data are means ± SD (n=3). **, p<0.01; Student’s t-test was used for statistical analysis. (D) ChIP-qPCR analysis showing the enrichment of OsNF-YB7 at the OsGLK1 promoter in 14-day-old OsNF-YB7-Flag seedling. Chromatin of each sample was immunoprecipitated using anti-Flag or IgG antibodies. Precipitated DNA was quantified by qPCR and DNA enrichment is displayed as a percentage of input DNA. Data are means ± SD (n=3). *, p<0.05; **, p<0.01; Student’s t-test was used for statistical analysis. ACTIN was used as a nonspecific target gene. The experiment was performed three times with similar results. The diagram in the upper panel showing the promoter structure of OsGLK1 and PCR amplicons (P1, P2, P3, P4, and P5) used for ChIP-qPCR. (E) Schematic diagram displaying the constructs used in the DLR assays of F. LUC, firefly luciferase; REN, Renilla luciferase; UAS, upstream activating sequence. (F) DLR assays showing that OsNF-YB7 represses the promoter activity of OsGLK1. Data are means ± SD (n=3). *, p<0.05; Student’s t-test was used for statistical analysis.
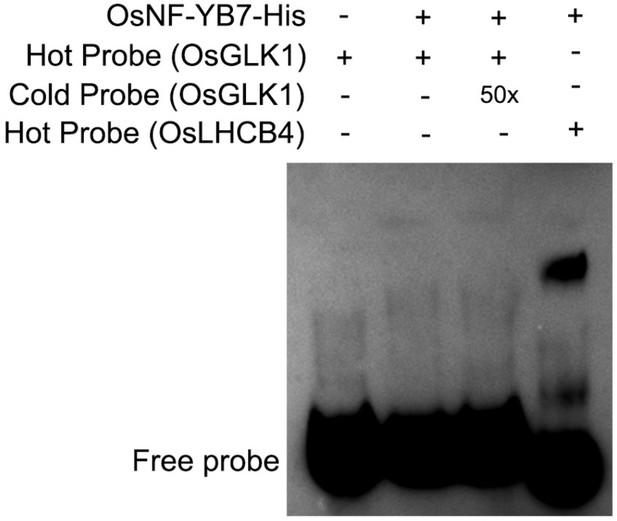
OsNF-YB7 does not directly binds to the promoter of OsGLK1 in vitro.
-
Figure 3—figure supplement 1—source data 1
Uncropped and labelled gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Raw unedited gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig3-figsupp1-data2-v1.zip
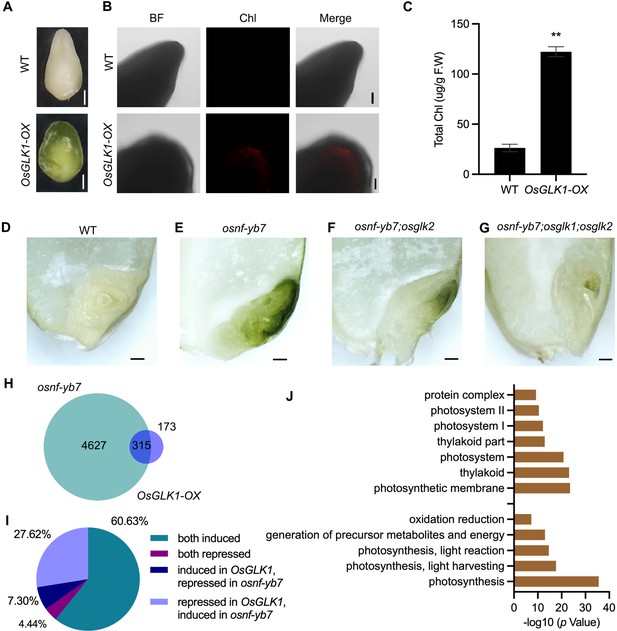
Chl biosynthesis in osnf-yb7 embryo requires active OsGLKs.
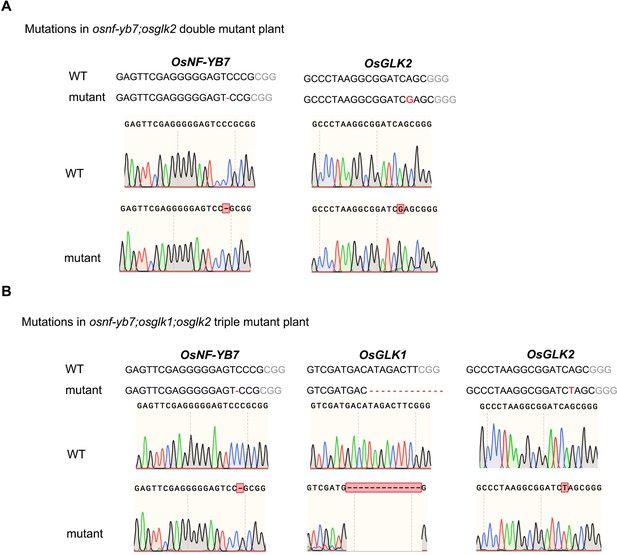
(A) Embryo morphologies of WT and OsGLK1-OX detached embryos at 10 DAF. Scale bars = 2 mm. (B) Chl autofluorescence of the WT and GLK-OX embryos at 10 DAF. BF, bright field; Chl, Chl autofluorescence. Scale bars = 100 μm. (C) Chl levels in WT and OsGLK1-OX embryos at maturation. Data are means ± SD (n=3). **, p<0.01; Student’s t-test was used for statistical analysis. (D–G) Morphologies of the embryos produced by WT (D), osnf-yb7 (E), osnf-yb7;osglk2 double mutant (F) and osnf-yb7;osglk1;osglk2 triple mutant (G). Scale bars = 1 mm. (H) A Venn diagram showing overlaps of the DEGs identified from the embryos of osnf-yb7 and OsGLK1-OX at 10 DAF. (I) A pie chart showing similar transcriptional changes of the common DEGs identified from osnf-yb7 and OsGLK1-OX. (J) Gene Ontology (GO) analysis of the common DEGs identified from osnf-yb7 and OsGLK1-OX.
-
Figure 4—source data 1
Differentially expressed genes (DEGs) in the 10-DAF-old embryos of OsGLK1-OX.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Common DEGs identified from the OsGLK1-OX and osnf-yb7 embryos at 10 DAF.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig4-data2-v1.xlsx
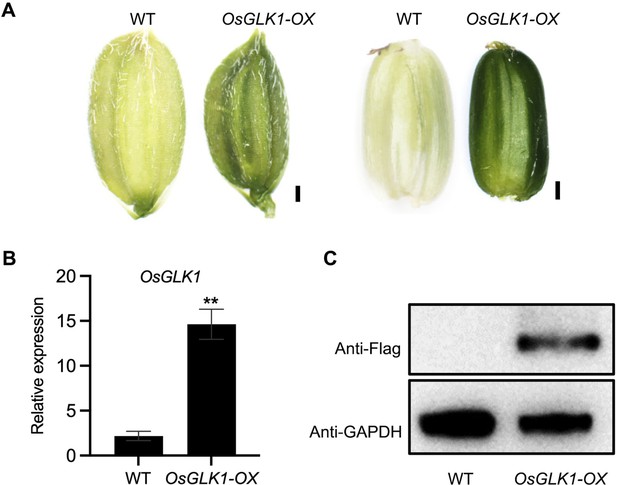
Overexpression of OsGLK1 induces chloroembryo in rice.
-
Figure 4—figure supplement 1—source data 1
Uncropped and labelled gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Raw unedited gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig4-figsupp1-data2-v1.zip
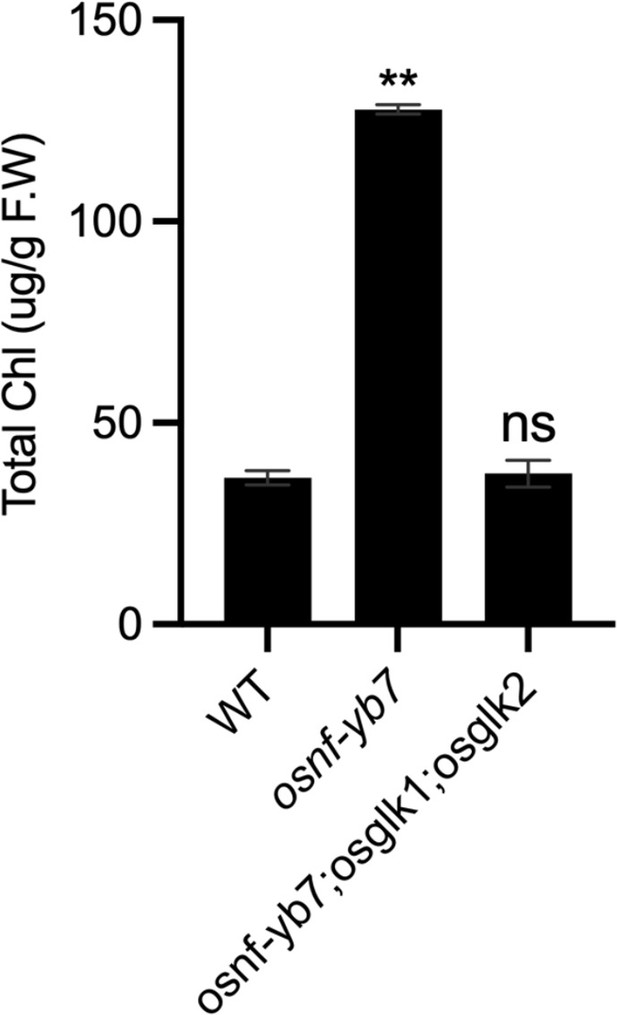
Chl levels in WT, osnf-yb7 and osnf-yb7;osglk1;osglk2 embryos at 10 DAF.
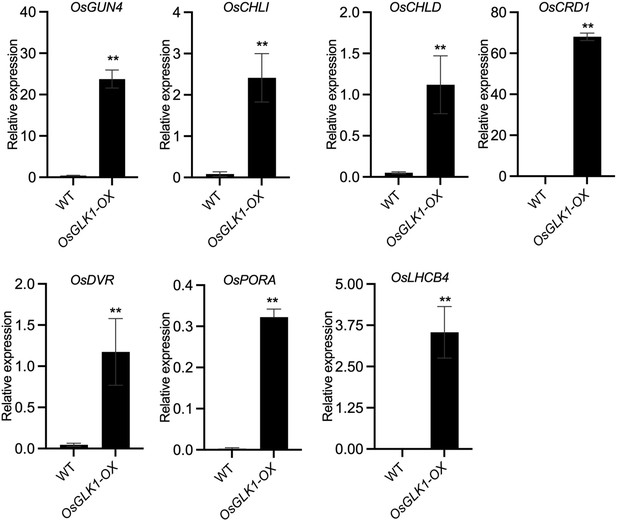
Overexpression of OsGLK1 activates the expression of Chl biosynthesis- and photosynthesis-related genes.
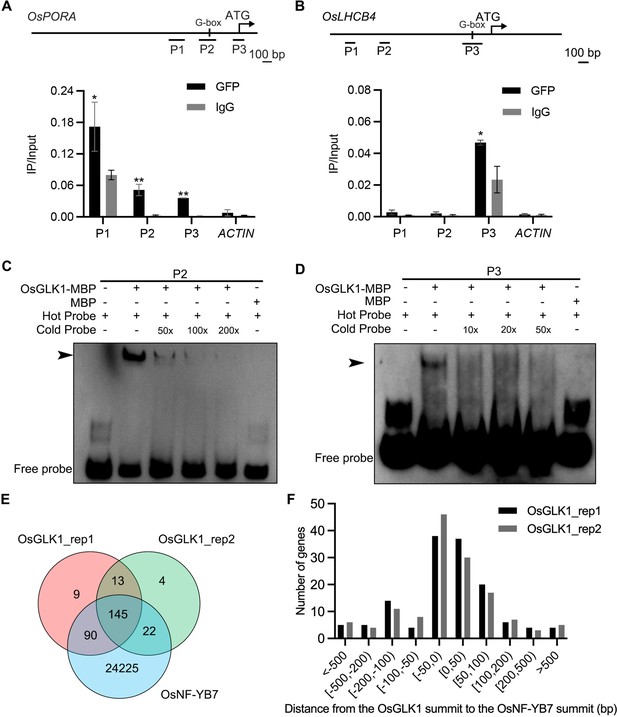
OsNF-YB7 and OsGLK1 regulate a common set of genes in the embryo.
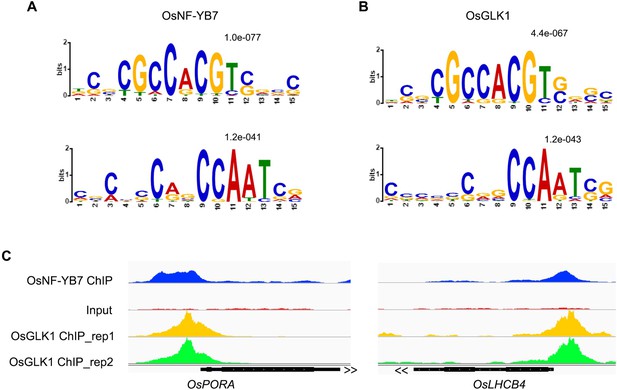
(A, B) ChIP-qPCR analysis showing the enrichment of OsGLK1 on the OsPORA (A) and OsLHCB4 (B) promoters. PCR amplicons used for ChIP-qPCR are indicated in the schematic diagrams. OsGLK1-GFP was transiently expressed in protoplasts isolated from green tissues of the 14-day-old WT seedling. Chromatin of each sample was immunoprecipitated using anti-GFP or igG antibodies. Precipitated DNA was quantified by qPCR and DNA enrichment is displayed as a percentage of input DNA. Data are means ± SD (n=3). *, p<0.05; **, p<0.01; Student’s t-test was used for statistical analysis. ACTIN was used as a nonspecific target gene. The experiment was performed three times with similar results. (C, D) EMSA assays showing that OsGLK1 directly binds to the promoters of OsPORA (C) and OsLHCB4 (D). The arrow heads indicate the shift bands. (E) A Venn diagram showing overlaps of the target genes between OsGLK1 and OsNF-YB7. (F) Distribution of the distance between the OsNF-YB7 and OsGLK1 summits showing that OsNF-YB7 and OsGLK1 bind to proximal regions of their common targets.
-
Figure 5—source data 1
Common targets of OsGLK1 and OsNF-YB7.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Uncropped and labelled gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig5-data2-v1.zip
-
Figure 5—source data 3
Raw unedited gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig5-data3-v1.zip
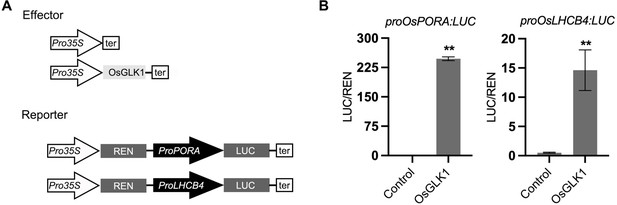
OsGLK1 significantly activates the promoter activities of OsPORA and OsLHCB4.
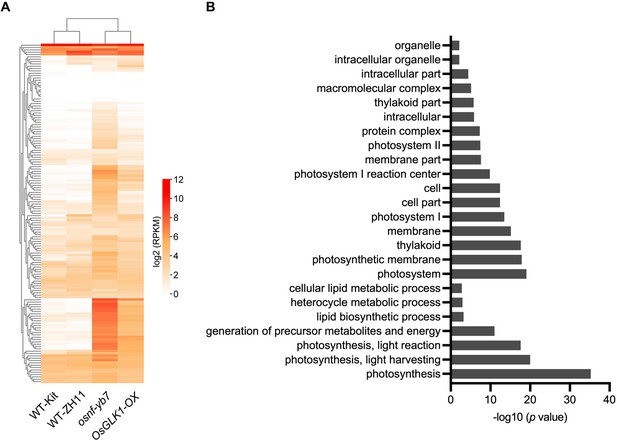
Many of the OsNF-YB7 and OsGLK1 common targets that activated in osnf-yb7 and OsGLK1-OX are involved in Chl biosynthesis and photosynthesis.
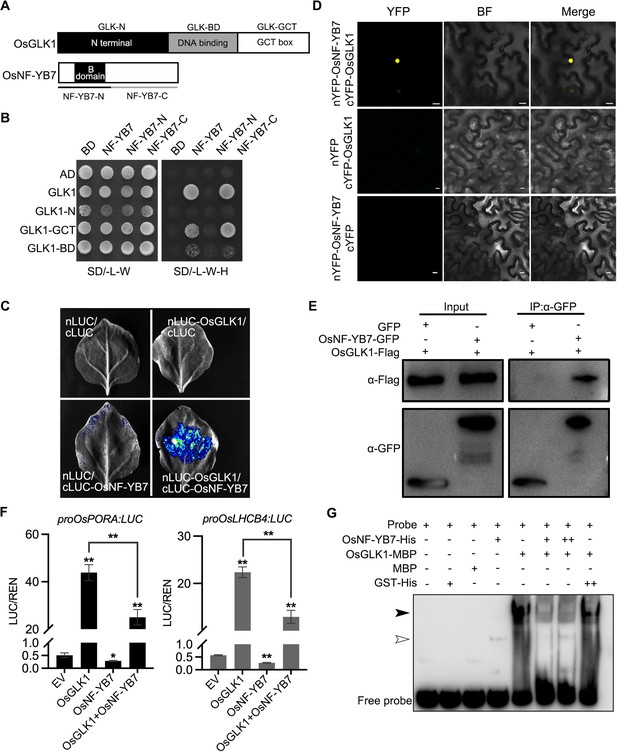
OsNF-YB7 interacts with OsGLK1 to regulate the expression of OsPORA and OsLHCB4.
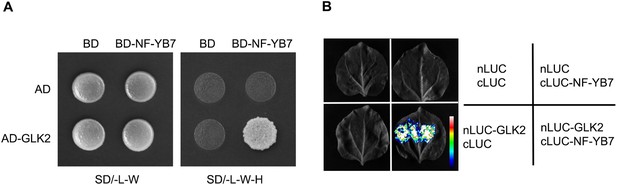
(A) Schematic diagrams showing the protein structures of OsGLK1 and OsNF-YB7. (B) Yeast-two-hybrid (Y2H) assays showing the interaction between OsNF-YB7 and OsGLK1. AD and BD indicate the activation domain and binding domain of GAL4, respectively. The full length or truncated OsNF-YB7 and OsGLK1 were fused with BD and AD, respectively. The indicated combinations of constructs were cotransformed into yeast cells and grown on the nonselective medium SD/-L-W and selective medium SD/-L-W-H. (C) A split complementary luciferase (LUC) assay confirmed the interaction between OsNF-YB7 and OsGLK1. Coexpression of the fusion of OsGLK1 and the N-terminal half of LUC (nLUC-OsGLK1) and the fusion of the C-terminal half of LUC and OsNF-YB7 (cLUC-OsNF-YB7) in the epidermal cells of N. benthamiana leaves induced LUC activities, whereas the epidermal cells coexpressing nLUC-OsGLK1 and cLUC, nLUC and cLUC-OsNF-YB7, or nLUC and cLUC did not show LUC activities. (D) Bimolecular fluorescence complementation (BiFC) assays showed interactions between OsNF-YB7 and OsGLK1 in the nucleus. OsNF-YB7 was fused with the N-terminal of yellow fluorescent protein (nYFP-OsNF-YB7); OsGLK1 was fused with the C-terminal of yellow fluorescent protein (cYFP-OsGLK1). The indicated combinations of constructs were transiently coexpressed in the leaf epidermal cells of N. benthamiana. Scale bar = 10 μm. (E) Co-immunoprecipitation (Co-IP) assays showing that OsNF-YB7 interacts with OsGLK1 in vivo. 35 S::OsNF-YB7:GFP (OsNF-YB7-GFP) and 35 S::OsGLK1:3×Flag (OsGLK1-Flag) were coexpressed in rice protoplasts and were immunoprecipitated with an anti-GFP antibody, and the immunoblots were probed with anti-GFP and anti-Flag antibodies. 35 S::GFP (GFP) was used as a negative control. (F) DLR assays showing that OsNF-YB7 represses the transactivation activity of OsGLK1 on OsPORA and OsLHCB4. Protoplasts isolated from etiolated seedlings were used for the analyses. EV, empty vector. Data are means ± SD (n=3). *, p<0.05; **, p<0.01; Student’s t-test was used for statistical analysis. (G) EMSA assay indicated that OsNF-YB7 inhibits the DNA binding of OsGLK1 to the promoter of OsPORA. The black and white arrow heads indicate the OsGLK1- and OsNF-YB7-bound probes, respectively; “+” and “++” indicate that 2- and 4 µM recombinant proteins were used for the reactions. The GST-His was used as a negative control.
-
Figure 6—source data 1
Uncropped and labelled gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig6-data1-v1.zip
-
Figure 6—source data 2
Raw unedited gels.
- https://cdn.elifesciences.org/articles/96553/elife-96553-fig6-data2-v1.zip
Additional files
-
Supplementary file 1
Primers used in this study.
- https://cdn.elifesciences.org/articles/96553/elife-96553-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96553/elife-96553-mdarchecklist1-v1.docx