Motor neurons are dispensable for the assembly of a sensorimotor circuit for gaze stabilization
Figures

phox2a loss-of-function mutants fail to develop nIII/nIV motor neurons and vertical eye rotation behavior.
Associated with Figure 1—figure supplement 2. (A) Schematic of vestibulo-ocular reflex circuitry and the genetic loss-of-function approach used to perturb motor-derived signals. (B) Fluorescent in situ hybridization showing phox2a transcript expression in statoacoustic ganglion sensory afferents (left), central projection neurons in the tangential nucleus (middle), or nIII/nIV extraocular motor neurons (right) at 5 days post-fertilization (dpf). Top: probe only, nuclei outlined with dashed lines. Bottom: probe (green) merged with somata, labeled by Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede) (sensory, central) or Tg(isl1:GFP) (motor). (C) Schematic of CRISPR/Cas9 mutagenesis approach. Top: Red star shows location of guides against phox2a DNA. Bottom: RNA sequence in wildtype and phox2ad22 alleles. Red dashed lines show deleted sequence; ‘STOP’ box shows predicted premature stop codon due to deletion. Right shows predicted protein sequence. (D) Transmitted light image of a 5 dpf wildtype (top) and phox2a null mutant (bottom). White arrows point to a normally inflated (top) or absent (bottom) swim bladder. (E) Images of nIII/nIV motor neurons in one hemisphere, labeled by Tg(isl1:GFP), in wildtype siblings (left) and phox2a null mutants (right) at 5 dpf. Scale bar, 20 µm. (F) Quantification of the number of Tg(isl1:GFP)+neurons in nIII/nIV from N=6 wildtype siblings and N=10 phox2a null mutants.
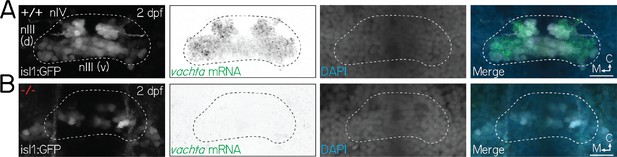
Extraocular motor neuron fate is perturbed in phox2a mutants.
(A) Fluorescent in situ hybridization images of nIII/nIV motor neurons, labeled by Tg(isl1:GFP), in wildtype larvae at 2 dpf. Panels from left to right show: (1) isl1+ motor neurons, (2) mRNA for vachta, a marker for cholinergic motor neurons, (3) DAPI, and (4) merge. White dashed lines show the extent of nIII/nIV. (B) Loss of both isl1 and vachta identity in nIII/nIV motor neurons in phox2a mutants at 2 dpf. All scale bars, 20 µm.
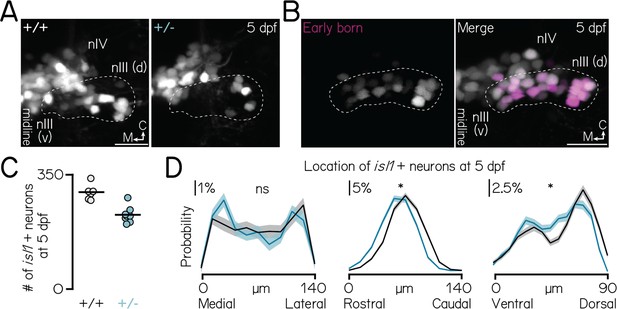
phox2a specifies nIII motor neuron fate in a dose- and birthdate-dependent manner.
(A) Images of nIII/nIV motor neurons, labeled in Tg(isl1:GFP), in wildtype siblings (left) and phox2a heterozygotes (middle) at 5 dpf. Wildtype image same as in Figure 1E. One hemisphere shown. White dashed lines outline the dorsal extent of nIII, which contains inferior rectus and medial rectus neurons (Greaney et al., 2017). Scale bar, 20 µm. (B) Location of the earliest-born neurons in nIII/nIV (left, magenta) against all nIII/nIV neurons labeled in Tg(isl1:Kaede) (right, grey). Larvae birthdated at 34 hpf (Materials and methods). One hemisphere shown. White dashed lines outline the dorsal extent of nIII. Scale bar, 20 µm. (C) Quantification of the number of Tg(isl1:GFP)+ neurons in nIII/nIV from N=6 wildtype siblings (grey) and N=8 phox2a heterozygotes (teal). Wildtype data same as Figure 1F. (D) Distributions showing probability of nIII/nIV soma location across each spatial axis in wildtype (black) and heterozygous (teal) phox2a larvae. Solid and shaded lines show mean and standard deviation, respectively, from bootstrapped data. Data from same fish quantified in C. ns, not significant; star, significant at the p<0.001 level.

Motor neurons are dispensable for proper connectivity between utricular sensory afferents and projection neurons.
Associated with Table 1. (A) Schematic of pitch vestibulo-ocular reflex circuitry. Dashed lines outline projection neurons as calcium imaging target. Nose-down/eyes-up channel represented with blue; orange, nose-up/eyes-down. (B) Schematic of tonic pitch-tilt stimulus delivered with Tilt-In-Place Microscopy (TIPM). Shaded regions show calcium imaging windows when fish were oriented horizontally immediately following tilts. Inset shows timecourse of the rapid step to restore horizontal position after tilts. Imaging experiments used larvae from the Tg(isl1:GFP);Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS:GCaMP6s) line. (C) Proportion of subtypes observed in sibling controls and phox2a null mutants. Blue: nose-down. Orange: nose-up. Grey: Neurons without directional tuning (criteria in Materials and methods). (D, E) Soma position of nose-down (blue) and nose-up (orange) neurons in sibling controls (left) and phox2a null mutants (right). Soma size scaled by strength of directional selectivity (min = 0; max = 1; see Materials and methods). (F/I) Heatmaps showing example tilt responses from nose-down (F) or nose-up (I) neurons in sibling controls (top) and phox2a null mutants (bottom). n=10 neurons with strongest ΔF/F responses to tilts shown. Each row shows an individual neuron. Shaded bars show calcium imaging window immediately following restoration from eccentric position. Black arrow points to first second of tilt response used for analyses. (G/J) Distributions of maximum ΔF/F responses to tilts for nose-down (G) or nose-up (J) neurons in sibling controls (black) and phox2a null mutants (red). Solid and shaded lines show mean and standard deviation, respectively, of bootstrapped data (Materials and methods). (H/K) Distributions of directional tuning score to tilts for nose-down (H) or nose-up (K) neurons in sibling controls (black) and phox2a null mutants (red). Tuning score ranges from 0 (equal responses to both tilt directions, no tuning) to 1 (responses to one tilt direction only); criteria detailed in Materials and methods. Solid and shaded lines show mean and standard deviation, respectively, of bootstrapped data.

Motor neurons are dispensable for proper connectivity between semicircular canal sensory afferents and projection neurons.
Associated with Table 1. (A) Schematic of impulse tilt experiments. Yellow dashed lines outline projection neurons as calcium imaging target. Impulse-responsive neurons (ventrally-localized) shown with purple; unresponsive neurons, grey. (B) Schematic of impulse stimuli delivered with TIPM. Shaded regions show calcium imaging windows at horizontal immediately following impulses. Inset shows timecourse of impulse stimulus. Imaging experiments used larvae from the Tg(isl1:GFP);Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS:GCaMP6s) line. (C) Proportion of impulse-responsive (purple) and unresponsive (grey) neurons observed in sibling controls and phox2a null mutants. (D) Soma position of impulse-responsive neurons in sibling controls (left) and phox2a null mutants (right). Soma size scaled by strength of calcium response (ΔF/F), normalized by max observed ΔF/F. (E) Heatmaps showing example impulse responses from neurons in sibling controls (left) and phox2a null mutants (right). n=10 example neurons shown. Each row shows an individual neuron. Shaded bars show calcium imaging window immediately following impulse delivery. Black arrow points to first second of tilt response used for analyses. Note smaller scale (0–0.75) of impulse responses relative to Figure 2F and I. (F) Distributions of maximum ΔF/F responses to impulses in sibling controls (black) and phox2a null mutants (red). Solid and shaded lines show mean and standard deviation, respectively, from bootstrapped data. (G) Distributions of directional tuning score to impulses in sibling controls (black) and phox2a null mutants (red). Tuning score ranges from 0 (equal responses to both tilt directions, no tuning) to 1 (responses to one tilt direction only); criteria detailed in Materials and methods. Solid and shaded lines show mean and standard deviation, respectively, from bootstrapped data.

Projection neurons are anatomically and molecularly poised to assemble with motor neuron partners in phox2a mutants.
(A) Schematic of retrograde photofill experiments. Projection neuron axons expressing the photolabile protein Kaede are targeted at the midbrain-hindbrain boundary with ultraviolet light. Converted protein (magenta) retrogradely diffuses to the soma, while the unconverted nucleus remains green. (B) Projection neuron somata in sibling controls (left) and phox2a null mutants (right) after retrograde photolabeling. Experiments performed at 5 dpf. Neurons visualized in Tg(isl1:GFP);Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS:E1b-Kaede). (C) Top two panels: Projection neuron axons at the hindbrain (inset) and midbrain-hindbrain boundary in sibling controls (top) and phox2a null mutants (bottom). Axons visualized using Tg(isl1:GFP);Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS:E1b-Kaede). White dashed outline (circle) shows arborization fields in nIII/nIV. Dashed box over axons shows location of two bottom panels. MHB and yellow dashed line, midbrain-hindbrain boundary. nucMLF: nucleus of the longitudinal fasciculus. Bottom two panels: Zoom of axons (dashed rectangle above). Spatial segregation between early-born (magenta +green) and late-born (green only) axons. White dashed line reflects separation between dorsal (nose-up, early-born) and ventral (nose-down, late-born) axon bundles. Image at 5 dpf in sagittal view. (D) Projection neuron axon bundle in a phox2a null mutant at 3 dpf. White arrows point to single collateral to two nIII/nIV neurons that were not eliminated following phox2a knockout. (E) Fluorescent in situ hybridization against RNA for three pre-synaptic markers: synaptophysin a (sypa; left), synaptic vesicle glycoprotein 2 (sv2, middle), and synapsin I (syn1, right). Top row, sibling controls. Bottom row, phox2a null mutants. For each panel set, left images show in situ probe expression (green) and right images show merge with projection neurons labeled in Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS:E1b-Kaede). Dashed lines outline the projection nucleus. Cell and transcript expression outside the projection nucleus is removed for visual clarity. Images taken at 5 dpf in sagittal mount. All scale bars, 20 µm.

Motor neurons are dispensable for normal transcriptional profiles of projection neurons.
Associated with Figure 5—figure supplement 1, Figure 5—figure supplement 4, Table 3. (A) Schematic of sequencing approach. Central projection neurons (Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS:E1b-Kaede)) are harvested from 3 dpf larvae. Flow cytometry is used to exclude neurons not labelled by Tg(–6.7Tru.Hcrtr2:GAL4-VP16). Bulk RNA sequencing is performed to compare the profiles of projection neurons in siblings and phox2a null mutants. (B) Example of projection neurons before (left) and after (right) harvesting. Neurons visualized with Tg(isl1:GFP);Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS:E1b-Kaede). Dashed lines outline projection neurons in the tangential nucleus; dotted lines, medial vestibular nucleus. Yellow region shows margin of harvesting error: non-projection neurons that may be included in bulk sequencing dataset. (C) Number of differentially expressed genes in projection neurons at 3 dpf after applying progressive filters based on gene expression in a reference single-cell dataset. Data shown on logarithmic scale. Solid, dashed, and dotted lines represent differentially-expressed gene with p adjusted<0.5, p adjusted<0.01, or p adjusted<0.001 significance, respectively. (D) Volcano plot showing differentially expressed genes in projection neurons between control and phox2a null larvae at 3 dpf. Dashed lines represent significance cutoffs: horizontal line, p >0.05; vertical line, Log2 Fold Change >2.0. Each circle is a gene. Genes to the left and right of 0 on the horizontal axis show downregulated and upregulated genes, respectively. Colors indicate percent of reference cells that express a given gene. Grey-colored genes are below both significance thresholds. (E) Same data as Figure 5D. Colored genes show eight candidates evaluated with fluorescent in situ hybridization: red, upregulated; blue, downregulated; yellow, highly-expressed controls (evx2). (F) Fluorescent in situ hybridization against candidate genes that met projection neuron filter criteria. Top row shows sibling controls; bottom row, phox2a null mutants. For each gene, left panels show RNA probe (green) and right panels show merge with projection neurons labeled by Tg(–6.7Tru.Hcrtr2:GAL4-VP16) (grey). Dashed lines outline the projection nucleus. Cell and transcript expression outside the projection nucleus is masked for visual clarity. Arrows denote whether genes are upregulated (red), downregulated (blue), or not significantly changed (yellow). Percentage refers to fraction of cells in a single-cell RNA sequencing reference atlas (Materials and methods) with detected transcript. Candidates: itga9 (log2 fold change = 23.0, p adj.=3.9 × 10–6), twf1b (log2 fold change = 5.9, p adj.=0.024), p4hb (log2 fold change = 5.1, p adj.=0.04), mapk6 (log2 fold change = 5.1, p adj.=0.06), rxfp2a (log2 fold change = −8.5, p adj.=1.1 × 10–5), satb1a (log2 fold change = −3.0, p adj.=0.001), evx2 (log2 fold change = 0.46, p adj.=0.99), myt1la (log2 fold change = 2.6, p adj.=0.44). All scale bars, 20 µm.
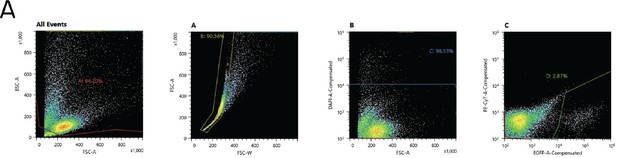
Flow cytometry gating strategy to sort fluorescently-labeled neurons for bulk RNA sequencing.
(A) Sequential gates used to sort fluorescent neurons labeled with Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede);Tg(isl1:GFP). Gate A excluded presumptive debris (small cells). Gate B isolated single cells and excluded large cells and doublets. Gate C excluded DAPI+ (dead or unhealthy) neurons. Gate D isolated fluorescent (GFP or Kaede+) neurons; neurons in this gate were sorted. Gates were set using negative controls (not shown; Materials and methods). Gates shown for one of four experimental repeats.
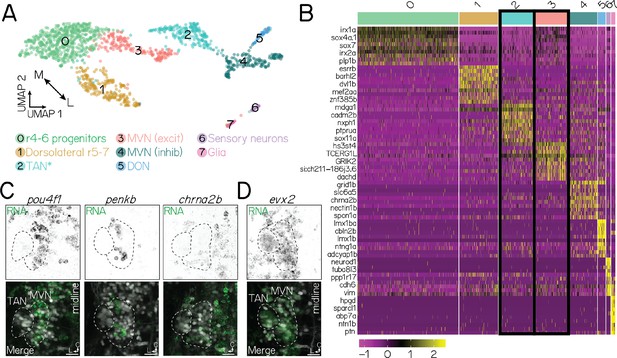
Molecular identification of projection neurons using a reference single-cell RNA sequencing atlas.
(A) UMAP visualization of a single-cell RNA sequencing atlas of n=1,468 neurons labeled in Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede), generated with 10 x Genomics (Materials and methods). Each circle is a single neuron. Neurons are clustered (colors) according to their transcriptional identity. Annotations are based on validated marker genes (data not shown). TAN, tangential nucleus; MVN, medial vestibular nucleus; r, rhombomere; DON, dorsal octavolateral nucleus. Note: Cluster 2 is broadly inclusive of lateral, excitatory vestibular projection neurons near the ventral base of the otic capsule. It includes, but is not exclusive to, the tangential nucleus. (B) Heatmap showing genes unique to each annotated cluster. Each row is a gene; names unlisted for clarity. Columns show distinct clusters. Color bar on top reflects clusters in A. Yellow and purple reflect stronger or weaker gene expression, respectively. Black outlines show two populations of interest: tangential nucleus neurons and medial vestibular nucleus neurons. (C) Fluorescent in situ hybridization against three markers (pou4f1, penkb, chrna2b) that are negative for tangential nucleus projection neurons and positive for medial vestibular nucleus neurons. Top row shows RNA expression (green); bottom row, merge with neurons labeled in Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede). Dashed lines outline the tangential nucleus (TAN) and medial vestibular nucleus (MVN). Data from 72 hpf larvae. Images shown in an axial view. (D) Fluorescent in situ hybridization against a positive marker (evx2) for both tangential nucleus and medial vestibular nucleus neurons. All scale bars, 20 µm.
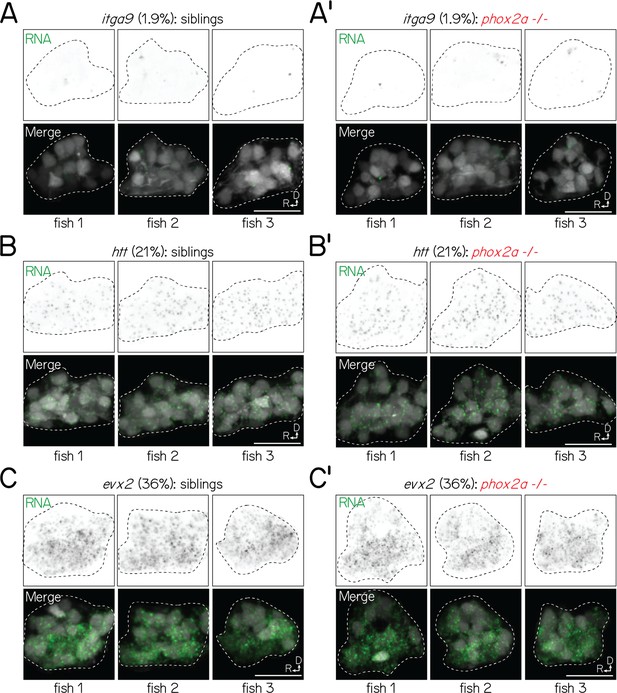
Visualization of transcripts in siblings and phox2a null mutants with fluorescent in situ hybridization is (1) consistent across larvae and (2) scales with predicted detection in projection neurons.
(A-A’) Fluorescent in situ hybridization against itga9 for three sibling (A) or phox2a null mutant (A’) larvae (72 hpf), imaged with identical conditions. Left column shows RNA (green); right column, merge with projection neurons visualized with Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede) (grey). Dashed lines outline the projection nucleus. Cell and transcript expression outside the projection nucleus is removed for visual clarity. Percentage (1.9%) refers to fraction of cells in a single-cell RNA sequencing reference atlas (Materials and methods) with detected transcript. All scale bars, 20 µm. (B-B’) Fluorescent in situ hybridization against htt for three sibling (B) and phox2a mutant (B’) larvae (72 hpf). (C-C’) Fluorescent in situ hybridization against evx2 for three sibling (C) and phox2a mutant (C’) larvae (72 hpf).
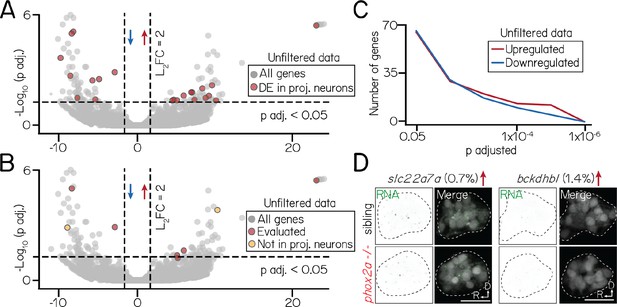
Differential gene expression in an unfiltered bulk sequencing dataset of siblings and phox2a mutants.
(A) Volcano plot showing differentially expressed genes across an unfiltered bulk RNA sequencing dataset. Dashed lines represent significance cutoffs: horizontal line, p adjusted>0.05; vertical line, Log2 Fold Change >2.0. Each circle is a gene. Genes to the left and right of 0 on the horizontal axis show downregulated and upregulated genes, respectively. Red color shows genes that are differentially expressed in a filtered subset of projection neurons (Figure 5). Grey-colored genes are below both significance thresholds. (B) Same data as A, now highlighting candidate genes evaluated by fluorescent in situ (Figure 5) with red. Two candidates (yellow) that did not meet projection neuron filter criteria (Materials and methods) are shown in D; remaining candidates (included in filtered data) shown in Figure 5F. (C) Same data as A-B, showing the number of differentially expressed genes at progressive significance thresholds (p adjusted). Red and blue lines show the number of significantly upregulated and downregulated genes, respectively. (D) Fluorescent in situ hybridization against two differentially expressed genes, slc22a7a (log2 fold change = 10.2, p adj.=1.6 × 10–4) and bckdhbl (log2 fold change = −9.1, p adj.=0.001), that did not meet projection neuron filter criteria. Percentage refers to fraction of all singly-profiled hindbrain vestibular neurons with expression (Materials and methods). Left columns show RNA (green); right columns, merge with projection neurons labeled with Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede) (grey). Dashed lines outline the projection nucleus. Cell and transcript expression outside the projection nucleus is removed for visual clarity. All scale bars, 20 µm.
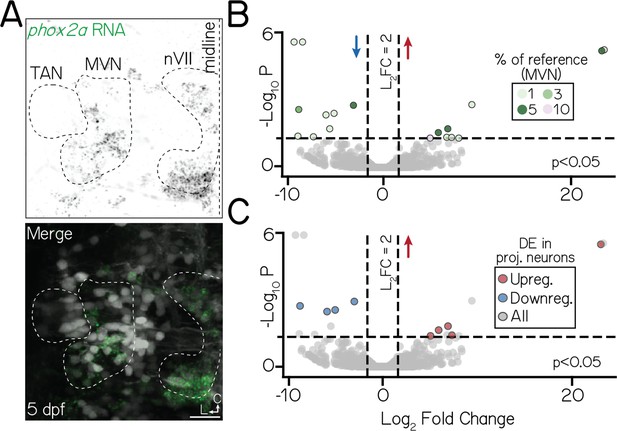
phox2a expression in the medial vestibular nucleus may underscore differential gene expression phenotypes in bulk data.
(A) Fluorescent in situ hybridization against phox2a in a 5 dpf larvae (axial view). Top panel shows phox2a RNA (green); bottom panel, merge with neurons visualized with Tg(isl1:GFP);Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede) (grey). White dashed lines outline three nuclei of interest: projection neurons in the tangential nucleus (TAN), the medial vestibular nucleus (MVN), and the facial nucleus (nVII). All scale bars, 20 µm. (B) Volanco plot showing differentially expressed genes in phox2a-expressing medial vestibular nucleus neurons between control and phox2a null larvae at 3 dpf. Dashed lines represent significance cutoffs: horizontal line, p>0.05; vertical line, Log2 Fold Change > 2.0. Each circle is a gene. Genes to the left and right of 0 on the horizontal axis show downregulated and upregulated genes, respectively. Colors indicate percent of reference phox2a-expressing medial vestibular neurons (Materials and methods, Figure 5—figure supplement 2A, B) that express a given gene. Grey-colored genes are below both significance thresholds. Data for all differentially expressed candidates in medial vestibular neurons shown in Table 6. (C) Same data as B. Color shows genes that are differentially expressed in both medial vestibular nucleus neurons and projection neurons.
Tables
Statistical comparisons of tilt responses across genotypes.
| WT (all) | WT (sampled) | phox2a+/+ | phox2a+/- | phox2a-/- | p val | |
|---|---|---|---|---|---|---|
| Tonic tilt stimuli | ||||||
| n (neurons/fish) | 255/10 | 125 /x | 76/5 | 109/6 | 297/16 | |
| % observed (nose-up/nose-down/untuned) | 50/44/7 | 37/54/9 | 40/54/7 | 56/37/7 | 44/50/6 | |
| ΔF/F, nose-up | 1.28 ± 1.23 | 1.27 ± 1.19 | 1.09 ± 1.03 | 1.12 ± 0.90 | 1.02 ± 0.82 | 0.26 |
| ΔF/F, nose-down | 2.01±1.66 | 1.99 ± 1.69 | 1.38 ± 0.91 | 1.98 ± 1.61 | 2.07±1.48 | 0.16 |
| directional tuning strength, nose-up | 0.84 ± 0.28 | 0.83 ± 0.30 | 0.87 ± 0.26 | 0.81 ± 0.28 | 0.81 ± 0.29 | 0.70 |
| directional tuning strength, nose-down | 0.72 ± 0.30 | 0.72 ± 0.31 | 0.73 ± 0.31 | 0.77 ± 0.30 | 0.68 ± 0.29 | 0.54 |
| Impulse stimuli | ||||||
| n (neurons/fish) | 255/10 | 125 /x | 76/5 | 109/6 | 297/16 | |
| % observed (responsive/unresponsive) | 58/42 | 57/43 | 57/43 | 60/39 | 70/30 | |
| ΔF/F | 0.41 ± 0.46 | 0.33 ± 0.28 | 0.29 ± 0.29 | 0.22 ± 0.16 | 0.32 ± 0.28 | 1.0E-05 |
| directional tuning strength | 0.08 ± 0.36 | 0.10 ± 0.38 | 0.003 ± 0.41 | 0.07 ± 0.48 | 0.07 ± 0.41 | 0.64 |
| Multiple comparisons | genotype | p val | Cohen’s d | |||
| ΔF/F to impulses | WT to sampled | p=0.13 | 0.21 | |||
| WT to +/+ | p=0.04 | 0.27 | ||||
| WT to +/- | p=3.8E-06 | 0.48 | ||||
| WT to -/- | p=0.006 | 0.24 | ||||
| +/+to +/- | p=0.47 | 0.34 | ||||
| +/+to -/- | p=0.89 | 0.11 | ||||
| +/-to -/- | p=0.02 | 0.49 |
Statistical comparisons of projection neuron topography across genotypes.
WT (sampled) refers to an n=125 neuron subset, sampled with replacement from a reference dataset of wildtype projection neurons. Data shown is the median/standard deviation distance from the ventro-lateral and rostral edges of the tangential nucleus (total size: 40 µm across each spatial axis). p val from one-way ANOVA (individual spatial axes) or multivariate ANOVA (global organization), respectively.
| WT (all) | WT (sampled) | phox2a+/+ | phox2a+/- | phox2a-/- | p val | |
|---|---|---|---|---|---|---|
| Tonic tilt stimuli | ||||||
| nose-down | ||||||
| n (cells/fish) | 111/10 | 51 /x | 41/5 | 40/6 | 147/16 | |
| dorsoventral | 20.0 ± 8.4 | 25.0 ± 8.4 | 15.0 ± 9.0 | 25.0 ± 8.7 | 20.0 ± 9.1 | 0.03 |
| mediolateral | 17.4 ± 6.0 | 17.6 ± 6.5 | 19.3 ± 6.6 | 15.3 ± 5.6 | 16.8 ± 6.6 | 0.15 |
| rostrocaudal | 16.8 ± 9.6 | 16.5 ± 9.6 | 19.5 ± 9.6 | 16.1 ± 10.4 | 18.2 ± 9.6 | 0.49 |
| global organization | 0.09 | |||||
| nose-up | ||||||
| n (cells/fish) | 111/10 | 51 /x | 41/5 | 40/6 | 147/16 | |
| dorsoventral | 30.0 ± 10.3 | 25.0 ± 10.8 | 27.5 ± 10.9 | 25.0 ± 10.4 | 30.0 ± 10.5 | 0.67 |
| mediolateral | 14.9 ± 7.1 | 15.1 ± 5.9 | 16.7 ± 7.0 | 18.6 ± 9.8 | 13.2 ± 7.2 | 9.6E-06 |
| rostrocaudal | 16.1 ± 9.8 | 16.7 ± 10.4 | 12.5 ± 10.7 | 14.8 ± 10.4 | 14.8 ± 10.7 | 0.51 |
| global organization | 2.7E-07 | |||||
| Multiple comparisons | genotype | p val | Cohen’s d | |||
| dorsoventral, nose-down | WT to sampled | 0.86 | 0.17 | |||
| WT to +/+ | 0.54 | 0.29 | ||||
| WT to +/- | 0.96 | 0.13 | ||||
| WT to -/- | 0.08 | 0.34 | ||||
| +/+to +/- | 0.35 | 0.41 | ||||
| +/+to -/- | 0.99 | 0.05 | ||||
| +/-to -/- | 0.07 | 0.45 | ||||
| mediolateral, nose-up | WT to sampled | 0.91 | 0.03 | |||
| WT to +/+ | 0.57 | 0.35 | ||||
| WT to +/- | 0.006 | 0.57 | ||||
| WT to -/- | 0.69 | 0.20 | ||||
| +/+to +/- | 0.74 | 0.23 | ||||
| +/+to -/- | 0.09 | 0.55 | ||||
| +/-to -/- | 5.4E-06 | 0.74 | ||||
| Impulse stimuli | ||||||
| n (responsive) | 148/10 | 138 /n | 43/5 | 55/5 | 214/16 | |
| dorsoventral | 20.0 ± 9.8 | 20.0 ± 10.1 | 20.0 ± 10.7 | 20.0 ± 10.9 | 20.0 ± 10.3 | 0.57 |
| mediolateral | 19.0 ± 5.9 | 16.1 ± 6.8 | 16.7 ± 8.6 | 20.4 ± 7.2 | 19.8 ± 5.8 | 0.003 |
| rostrocaudal | 21.9 ± 9.9 | 17.3 ± 9.8 | 15.4 ± 9.1 | 13.0 ± 9.9 | 22.0 ± 9.5 | 2.9E-05 |
| global organization | 1.2E-0.9 | |||||
| Multiple comparisons | genotype | p val | Cohen’s d | |||
| mediolateral, responsive | WT to sampled | 0.99 | 0.002 | |||
| WT to +/+ | 0.32 | 0.37 | ||||
| WT to +/- | 9.82 | 0.17 | ||||
| WT to -/- | 0.33 | 0.23 | ||||
| +/+to +/- | 0.91 | 0.14 | ||||
| +/+to -/- | 0.007 | 0.55 | ||||
| +/-to -/- | 0.05 | 0.37 | ||||
| rostrocaudal, responsive | WT to sampled | 0.98 | 0.37 | |||
| WT to +/+ | 0.12 | 0.29 | ||||
| WT to +/- | 0.001 | 0.53 | ||||
| WT to -/- | 0.07 | 0.10 | ||||
| +/+to +/- | 0.90 | 0.72 | ||||
| +/+to -/- | 0.95 | 0.33 | ||||
| +/-to -/- | 0.23 | 0.21 |
Differentially expressed genes in projection neurons.
Star indicates a gene was evaluated using fluorescent in situ hybridization. # symbol indicates a gene was also differentially expressed in adjacent phox2a-expressing medial vestibular neurons (see Figure 5—figure supplement 5). ”% of projection neurons with expression” refers to detection in a filtered subset of projection neurons from a single-cell reference atlas of neurons labeled in Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede) (Materials and methods, Figure 5—figure supplement 2). Genes sorted by p adjusted value. Data associated with Figure 5.
| Gene | % of projection neurons with expression | Log2 fold change | p adjusted |
|---|---|---|---|
| Upregulated | |||
| * # itga9 | 1.9 | 23.0 | 3.9E-06 |
| si:dkey-54n8.2 | 1.3 | 9.6 | 0.007 |
| myof | 1.3 | –8.3 | 0.010 |
| gbe1a | 1.9 | 7.0 | 0.01 |
| # dysf | 1.9 | 6.9 | 0.016 |
| * # twf1b | 2.5 | 5.9 | 0.024 |
| cers3a | 2.5 | 9.2 | 0.025 |
| asip2b | 1.3 | 8.7 | 0.032 |
| # pole | 1.3 | 7.3 | 0.041 |
| abtb2a | 3.1 | 4.7 | 0.041 |
| * # p4hb | 6.3 | 5.1 | 0.044 |
| postnb | 3.8 | 10.1 | 0.044 |
| fhdc3 | 2.5 | 4.5 | 0.044 |
| Downregulated | |||
| msmo1 | 1.3 | –8.3 | 8.4E-06 |
| *rxfp2a | 4.4 | –8.5 | 1.1E-05 |
| si:ch73-204p21.2 | 1.3 | –9.9 | 2.2E-04 |
| tsta3 | 0.0 | –6.6 | 6.8E-04 |
| * # satb1a | 8.2 | –3.0 | 0.001 |
| # polrmt | 1.9 | –8.7 | 0.002 |
| # znf975 | 1.3 | –5.0 | 0.003 |
| # phldb1a | 3.1 | –5.8 | 0.004 |
| asns | 2.5 | –7.8 | 0.032 |
| nr1i2 | 1.9 | –5.5 | 0.039 |
| Control | |||
| evx2 | 36.4 | 0.46 | 0.99 |
| myt1la | 80.5 | 2.6 | 0.44 |
Top 50 expressed genes in an unfiltered bulk RNA sequencing dataset of phox2a siblings.
‘% of unfiltered 10 x neurons’ refers to gene detection in a single-cell atlas of neurons labeled in Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede) (n=1,468 neurons). ‘% of projection neurons’ refers to gene detection in a subset of the single-cell atlas containing projection neurons in the tangential nucleus (n=159 neurons). Data associated with Figure 5.
| Gene | % of unfiltered 10 x neurons with expression | % of projection neurons |
|---|---|---|
| ints5 | 23.2 | 15.1 |
| stmn1b | 78.6 | 78.6 |
| sox4a | 6.5 | 3.1 |
| basp1 | 61.4 | 62.9 |
| hmgb3a | 68.2 | 69.8 |
| ptmaa | 84.5 | 77.4 |
| gapdhs | 28.9 | 30.2 |
| pnrc2 | 81.3 | 78.6 |
| snap25a | 65.7 | 78.0 |
| gpm6ab | 81.1 | 78.6 |
| calm3b | 0.0 | 0.0 |
| marcksl1b | 88.8 | 86.2 |
| tuba1c | 59.9 | 54.7 |
| cd81a | 43.3 | 49.7 |
| meis1b | 87.9 | 87.4 |
| rtn1a | 73.4 | 74.8 |
| elavl3 | 87.1 | 82.4 |
| hmgb1b | 57.2 | 49.7 |
| ptmab | 80.7 | 73.6 |
| zc4h2 | 56.9 | 58.5 |
| meis2b | 57.1 | 49.7 |
| slc25a5 | 51.4 | 55.3 |
| mab21l2 | 62.7 | 68.6 |
| h3f3c | 69.1 | 61.6 |
| rtn1b | 36.4 | 33.3 |
| elavl4 | 78.7 | 67.3 |
| gng3 | 37.2 | 42.8 |
| pik3r3b | 77.4 | 83.6 |
| tubb5 | 25.3 | 25.2 |
| histh1l | 61.0 | 62.9 |
| serinc1 | 51.9 | 59.1 |
| ckbb | 23.5 | 30.8 |
| az1a | 43.5 | 49.7 |
| az1b | 36.9 | 38.4 |
| actb1 | 23.6 | 28.3 |
| ywhaba | 36.2 | 40.9 |
| ywhag2 | 36.2 | 50.9 |
| si:ch211-222l21.1 | 73.8 | 63.5 |
| si:dkey-276j7.1 | 45.8 | 56.0 |
| aldocb | 19.3 | 17.6 |
| actb2 | 27.0 | 30.8 |
| tmem59l | 39.8 | 56.6 |
| calm2b | 37.9 | 47.2 |
| hmgn6 | 73.6 | 65.4 |
| h2afx1 | 59.6 | 53.5 |
| cd99l2 | 32.4 | 36.5 |
| cirbpb | 77.8 | 73.6 |
| ppdpfb | 74.5 | 65.4 |
| stxbp1a | 52.3 | 66.0 |
| Control | ||
| evx2 | 33.8 | 36.4 |
Top 50 differentially expressed genes in an unfiltered bulk RNA sequencing dataset of phox2a siblings and null mutants.
One star indicates a gene was retained in a filtered subset of projection neurons; %, evaluated using fluorescent in situ hybridization. ‘% of unfiltered 10 x neurons’ refers to gene detection in an unfiltered single-cell reference atlas of neurons labeled in Tg(–6.7Tru.Hcrtr2:GAL4-VP16);Tg(UAS-E1b:Kaede) (n=1,468 neurons). Putative origin inferred from gene expression in the annotated 10 x dataset (Materials and methods, Figure 5—figure supplement 2). Genes sorted by p adjusted value. Data associated with Figure 5.
| Gene | % of unfiltered 10 x neurons with expression | Putative origin | Log2 fold change | p adjusted |
|---|---|---|---|---|
| Upregulated | ||||
| macc1 | 0.1 | r4-6 | 24.0 | 3.2E-06 |
| CR559941.1 | 0.0 | 23.7 | 3.4E-06 | |
| si:dkey-65b12.6 | 0.0 | 23.5 | 3.4E-06 | |
| si:ch73-106n3.2 | 0.1 | 23.5 | 3.4E-06 | |
| mcm10 | 0.1 | MNs | 23.4 | 3.4E-06 |
| si:ch211-244o22.2 | 0.5 | r4-6 | 23.4 | 3.4E-06 |
| dre-mir-10a | 0.0 | 23.3 | 3.5E-06 | |
| itga4 | 0.3 | r5-6 (inhibitory) | 23.2 | 3.5E-06 |
| si:dkeyp-87d8.8 | 0.0 | 23.2 | 3.6E-06 | |
| arsj | 0.5 | MNs | 23.0 | 3.9E-06 |
| tlr1 | 0.0 | 23.0 | 3.9E-06 | |
| * % itga9 | 2.3 | r4-7 | 23.0 | 3.9E-06 |
| tofb | 0.5 | r4-6 | 9.1 | 1.4E-05 |
| myo7ba | 0.4 | r4-7 | 9.8 | 1.0E-04 |
| zfand1 | 0.1 | MNs | 9.0 | 1.5E-04 |
| % slc22a7a | 0.7 | r4-7 | 10.2 | 1.5E-04 |
| agrp | 0.0 | 13.4 | 4.1E-04 | |
| si:dkey-46i9.6 | 0.1 | r5-7 | 7.7 | 6.8E-04 |
| muc2.2 | 0.0 | 9.4 | 6.9E-04 | |
| cd37 | 0.0 | 9.1 | 9.8E-04 | |
| musk | 0.3 | r4-6 | 9.4 | 1.2E-03 |
| mcamb | 0.2 | r5-7 | 8.3 | 2.7E-03 |
| ppp1r42 | 0.5 | r5-6 (inhibitory) | 7.9 | 3.1E-03 |
| CR677513.1 | 0.0 | 9.9 | 3.5E-03 | |
| Downregulated | ||||
| * % satb1a | 7.9 | r4-7 (inc inhib), MNs | –8.6 | 1.0E-06 |
| *znf975 | 0.7 | r4-6 | –8.3 | 1.5E-06 |
| phldb1a | 0.6 | r5-7 (inc inhib) | –9.2 | 1.5E-06 |
| TSTA3 | 0.0 | –9.7 | 3.4E-06 | |
| si:dkey-24p1.6 | 0.0 | –8.3 | 8.4E-06 | |
| si:dkey-77f5.14 | 0.2 | r5-7 | –8.5 | 1.1E-05 |
| tha1 | 0.1 | MVN | –10.3 | 2.1E-05 |
| serpinh2 | 0.5 | r4-6 | –9.0 | 3.7E-05 |
| ghrh | 0.3 | r4-7 | –9.5 | 6.9E-05 |
| asah1b | 0.8 | r4-7 | –7.8 | 9.9E-05 |
| msmo1 | 0.9 | r5-7, inc inhib | –8.9 | 1.1E-04 |
| tagln2 | 0.3 | glia | –8.4 | 2.2E-04 |
| zgc:174863 | 0.1 | MNs | –9.9 | 2.2E-04 |
| * % rxfp2a | 3.2 | r4-7, inc inhib | –6.6 | 6.8E-04 |
| bmp4 | 0.7 | r4-7 | –6.8 | 6.8E-04 |
| cfl1l | 0.1 | r4-6 | –8.4 | 6.8E-04 |
| * polrmt | 4.2 | r4-7, inc inhib | –8.8 | 6.9E-04 |
| anxa2a | 0.6 | r4-7 | –3.0 | 1.3E-03 |
| galr1a | 0.3 | MVN | –9.1 | 1.4E-03 |
| selenow2b | 0.1 | –8.0 | 1.8E-03 | |
| % bckdhbl | 1.4 | r4-7, glia, MNs | –8.7 | 2.1E-03 |
| boka | 0.5 | r5-7 | –8.6 | 2.9E-03 |
| cyldb | 0.2 | r4-7 | –7.9 | 3.0E-03 |
| pon2 | 0.6 | r4-7, glia, MNs | –5.0 | 3.1E-03 |
| si:ch73-204p21.2 | 0.3 | r5-7, inc inhib | –8.2 | 3.5E-03 |
| and2 | 0.1 | r4-6 | –5.8 | 3.7E-03 |
| Control | ||||
| evx2 | 33.8 | r4-7 | 0.46 | 0.99 |
Differentially expressed genes in phox2a-expressing medial vestibular neurons.
Star indicates a gene was evaluated in projection neurons using fluorescent in situ hybridization. # indicates a gene was significantly differentially expressed in projection neurons. ‘% of medial vestibular neurons’ refers to detection in a subset of phox2a-expressing medial vestibular neurons in a single-cell reference atlas (Materials and methods, Figure 5—figure supplement 2). ‘% of projection neurons with expression’ refers to detection in a filtered subset of projection neurons. Gene sorted by p adjusted value. Data associated with Figure 5.
| Gene | % of medial vestibular neurons | % of projection neurons | Log2 fold change | p adjusted |
|---|---|---|---|---|
| Upregulated | ||||
| itga4 | 2.2 | 0.6 | 23.2 | 3.5E-06 |
| * # itga9 | 6.7 | 1.9 | 23.0 | 3.9E-06 |
| musk | 2.2 | 0.6 | 9.4 | 0.001 |
| # dysf | 6.7 | 1.9 | 6.9 | 0.016 |
| * # twf1b | 8.9 | 1.9 | 5.9 | 0.024 |
| gabrr2a | 2.2 | 2.5 | 6.8 | 0.040 |
| # pole | 2.2 | 0.0 | 7.3 | 0.041 |
| * # p4hb | 15.6 | 1.3 | 5.1 | 0.044 |
| col27a1b | 2.2 | 0.6 | 8.0 | 0.044 |
| Downregulated | ||||
| asah1b | 2.2 | 0.0 | –8.3 | 1.5E-06 |
| boka | 2.2 | 0.0 | –9.2 | 1.5E-06 |
| * # satb1a | 6.7 | 8.2 | –3.0 | 0.001 |
| # polrmt | 4.4 | 1.9 | –8.7 | 0.002 |
| # znf975 | 2.2 | 1.3 | –5.0 | 0.003 |
| # phldb1a | 2.2 | 3.1 | –5.8 | 0.004 |
| fosl1a | 2.2 | 0.0 | –5.4 | 0.016 |
| pitpnaa | 2.2 | 0.0 | –8.8 | 0.036 |
| sgpp1 | 2.2 | 0.6 | –7.1 | 0.038 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Tween | Thermo Fisher Scientific | BP337-100 | |
| Chemical compound, drug | 32% paraformaldehyde | Electron Microscopy Sciences | 15714 | |
| Chemical compound, drug | Proteinase K | Thermo Fisher Scientific | 25530049 | |
| Peptide, recombinant protein | Papain | Worthington Biochemical | LK003178 | |
| Chemical compound, drug | Hanks Buffered Salt Solution (HBSS) | Thermo Fisher Scientific | 14170112 | |
| Chemical compound, drug | Earl’s Buffered Salt Solution (EBSS) | Thermo Fisher Scientific | 24010043 | |
| Peptide, recombinant protein | DNAse | Worthington Biochemical | LK003172 | |
| Peptide, recombinant protein | DAPI | Invitrogen | D1306 | |
| Chemical compound, drug | L15 Medium | Thermo Fisher Scientific | 11415064 | |
| Chemical compound, drug | Fetal bovine serum, qualified, triple-filtered | Thermo Fisher Scientific | A3160501 | |
| Peptide, recombinant protein | Collagenase Type 1 A | Sigma Aldrich | C9891-500MG | |
| Chemical compound, drug | Low melting point agarose | Thermo Fisher Scientific | 16520 | |
| Chemical compound, drug | Ethyl-3-aminobenzoic acid ethyl ester (MESAB) | Sigma Aldrich | E10521 | |
| Chemical compound, drug | Pancuronium bromide | Sigma Aldrich | P1918 | |
| Commercial assay, kit | in situ hybridization chain reaction v3.0 (HCR) | Molecular Instruments | N/A | |
| Commercial assay, kit | RNAqueous Micro Total RNA Isolation Kit | Thermo Fisher Scientific | AM1931 | |
| Commercial assay, kit | MEGAshortscript T7 Transcription Kit | Thermo Fisher Scientific | AM1354 | |
| Commercial assay, kit | QiaQUICK PCR Purification Kit | Qiagen | 28104 | |
| Commercial assay, kit | EnGen Spy Cas9 NLS | New England Biolabs | M0646T | |
| Strain, strain background (Danio rerio) | Tg(–6.7Tru.Hcrtr2:GAL4-VP16) | Lacoste et al., 2015; Schoppik et al., 2017 | ZFIN: ZDB-TGCONSTRCT-151028–8 | |
| Strain, strain background (Danio rerio) | Tg(UAS-E1b:Kaede) | Scott et al., 2007 | ZFIN: ZDB-TGCONSTRCT-070314–1 | |
| Strain, strain background (Danio rerio) | Tg(isl1:GFP) | Higashijima et al., 2000 | ZFIN: ZDB-ALT-030919–2 | |
| Strain, strain background (Danio rerio) | Tg(UAS:GCaMP6s) | Chen et al., 2013 | ZFIN: ZDB-TGCONSTRCT-140811–3 | |
| Strain, strain background (Danio rerio) | phox2ad22 | This study | N/A | |
| Strain, strain background (Danio rerio) | phox2ad19 | This study | N/A | |
| Strain, strain background (Danio rerio) | phox2ai2 | This study | N/A | |
| Sequence-based reagent (primers) | phox2a forward primer | Sigma Aldrich | N/A | CAGCCAGAGCAACGGCTTCC |
| Sequence-based reagent (primers) | phox2a reverse primer | Sigma Aldrich | N/A | AAGCCGACAACAGTGTGTGTGTAA |
| Sequence-based reagent (primers) | phox2a guide 1 | Sigma Aldrich | N/A | CTCGCCACCGCCAGCTGCAC |
| Sequence-based reagent (primers) | phox2a guide 2 | Sigma Aldrich | N/A | CTCCGGCTTCAGCTCCGGCC |
| Sequence-based reagent (oligonucleotides) | HCR probes | Integrated DNA Technologies | N/A | |
| Software, algorithm | Fiji/ImageJ | Schindelin et al., 2012 | RRID: SCR_02285 | |
| Software, algorithm | Adobe Illustrator (2021) | Adobe | RRID: SCR_010279 | |
| Software, algorithm | Matlab 2020b | Mathworks | RRID: SCR_001622 | |
| Software, algorithm | Seurat v4 | Hao et al., 2021 | https://satijalab.org/seurat | |
| Software, algorithm | CRISPR Guide RNA Design Tool | Benchling | https://benchling.com/crispr | |
| Other | 20 micron cell strainer | pluriSelect | 431002060 | Method details (Neuron harvesting) |
| Other | SH800z 100 micron sorting chip | Sony | LE-C3210 | Method details (Flow cytometry) |





