mirror determines the far posterior domain in butterfly wings
Figures
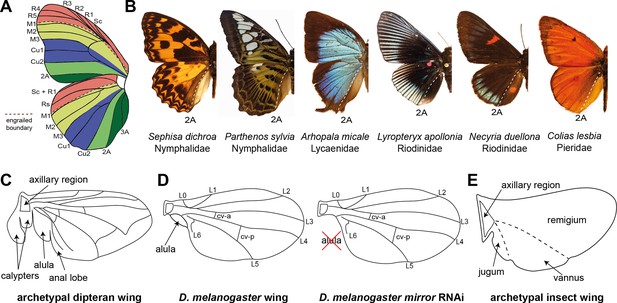
Proposed anteroposterior (AP) domains of insect wings.
(A) Comparative morphology previously identified boundaries associated with color pattern variation, demarcated by the M1, M3, Cu2, and 2A veins (de Celis et al., 1996). (B) Examples of the 2A vein marking a clear posterior color pattern boundary across multiple butterfly families. (C) Proximal posterior features of the archetypal dipteran wing include the calypters, which are only found in the Calyptratae clade, and the alula, a lobe at the base of the wing blade (McAlpine et al., 1981). (D) mirror RNAi knockdowns result in highly specific loss of the alula in Drosophila melanogaster (Kehl et al., 1998). (E) Snodgrass’ model of the archetypal insect wing specifies three major domains along the anteroposterior axis: the remigium, the vannus, and the jugum. Homologies of these domains with the features of the dipteran wing (C), or butterfly wing pattern boundaries (A), have remained unclear.
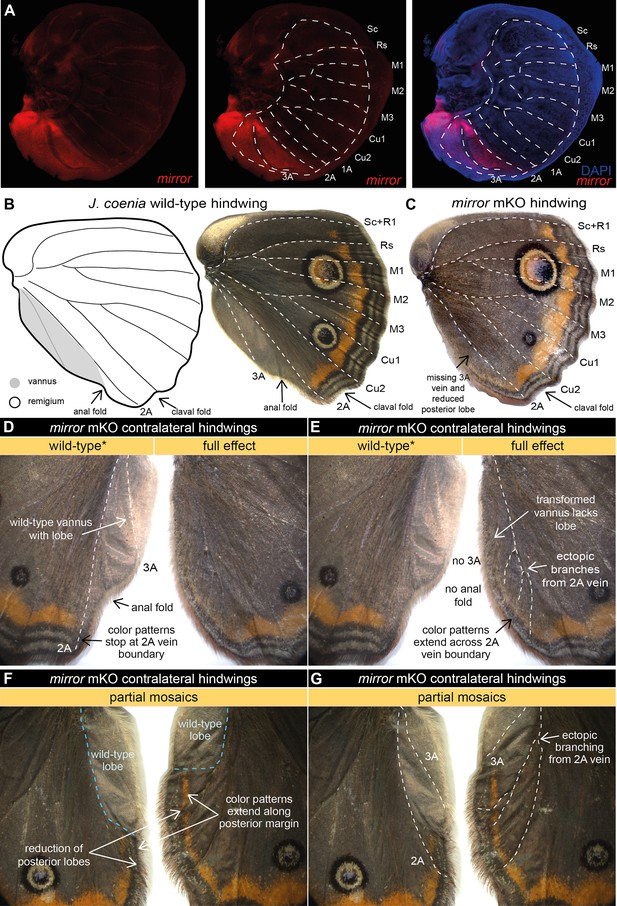
mirror determines the identity of the far posterior vannus domain in J. coenia.
(A) In J. coenia, mirror (red) is expressed posterior to the 2A vein in the late last-instar hindwing imaginal discs, marking the region that will develop into the vannus. See Figure 2—figure supplement 1 for forewing expression and controls. (B) In adult butterfly wings, the vannus is the wing field posterior to the 2A vein, shown here in J. coenia as a posterior lobe with silvery scales devoid of color patterns found anterior to 2A. (C) Targeted mosaic knockouts (mKOs) of mirror result in loss of vannus features in J. coenia. (D) Annotation of wild-type features preserved on one side of a contralateral mKO. (E) Annotation of full effect mutant phenotype in the opposing wing from the individual in (D) highlights loss of lobe, loss of 3A, ectopic branching of 2A, and posterior extension of color patterns. (F) Annotation of a partial mKO highlights transformation of posterior lobe and extension of color patterns, both of which terminate at the mutant clonal boundary. (G) The individual shown in (F) also shows ectopic branching of 2A in the mutant region, but preservation of 3A in the wild-type region. For additional mirror mKO phenotypes, see Figure 2—figure supplements 2–4.
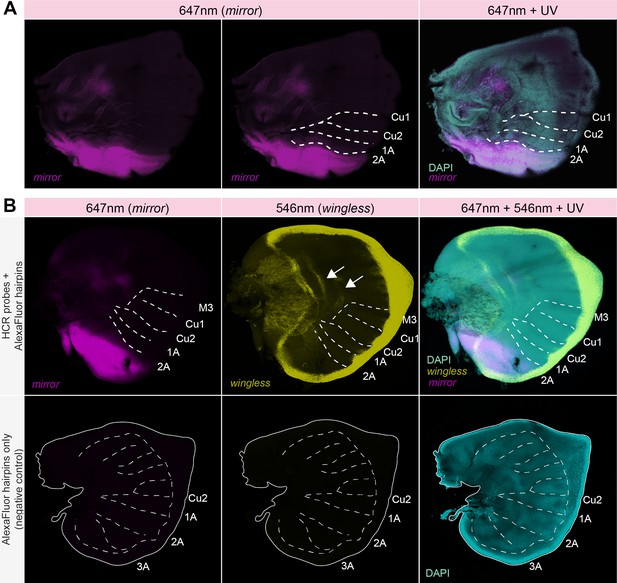
Hybridization chain reaction (HCR) in situ hybridization of mirror highlights posterior domain expression in J. coenia wing imaginal discs.
Related to Figure 2A. (A) mirror is expressed posterior to the 2A vein in last-instar forewing imaginal discs of J. coenia. (B) Double in situ of mirror (Channel 1: 647 nm) and wingless (Channel 2: 546 nm) in last instar J. coenia hindwing imaginal discs. Expression of wingless observed by HCR precisely matches expression in wing margin peripheral tissue and discal bands (white arrow) as previously described from chromogenic in situ (de Celis et al., 1996), thus providing a positive control for our HCR protocol. Bottom panels depict negative controls, lacking HCR probes, that show no signal in the 647 nm or 546 nm channels. Dashed lines show the position of selected veins.

Mosaic knockouts (mKOs) of mirror result in partial or entire loss of posterior wing domain identity in dorsal J. coenia hindwings.
Related to Figure 2B–E. (A) Comparison of wild-type dorsal hindwing to mutant wings reveals complete or partial transformation of the vannus (black arrows), expansion of color patterns into the posterior wing margin (white arrows) along with vein aberrations (denoted by *), or total loss (denoted by -). The middle panels show magnified images of the posterior wing margin and vannus, with scale morphology details, in wild-type and mKO mutants (complete transformation in JCM22 and JCM29; partial transformation in JCM41). The bottom panels show contralateral hindwing pairs from the same individuals showing asymmetry in clonal mutations affecting posterior wing margin phenotype. (B) Additional mirror mKO hindwings show vannus transformation phenotypes, annotated as described above.

Mosaic knockouts (mKOs) of mirror result in partial or entire transformation of the posterior wing domain.
Related to Figure 2B–E. mirror mKOs in ventral J. coenia hindwings resulting in the expansion of color pattern elements (white arrows) as seen in asymmetric contralateral pairs from the same mutant individuals. Abnormal venation annotated by (*), missing venation annotated by (-).

mirror mosaic knockouts (mKOs) show posterior margin anomalies in J. coenia forewings.
mKOs of mirror result in anomalies along the forewing 2A vein, including vein bifurcation and ectopic vein fragments, which are often associated with color pattern disruption. Dashed lines show the position of 2A. Black arrows highlight 2A abnormalities.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Junonia coenia) | mirror | This paper | V2 genome: JC_02269-RA; V3 genome: JC_g12219 | V2 genome is available on https://www.lepbase.org/. Email authors if http://lepbase.org/ is inaccessible. |
| Strain (Junonia coenia) | Lab based for many generations – derived from wild collected individuals | |||
| Sequence-based reagent | mirror HCR v3 probes | This paper; Molecular Instruments | Lot #PRQ901 | |
| Sequence-based reagent | wingless HCR v3 probes | Molecular Instruments | Lot #PRG129 | |
| Sequence-based reagent | mirror sgRNA-1 | This paper; IDT | CRISPR-cas9 guide RNA | 5’-GAATGGACTTGAACGGGGCA |
| Sequence-based reagent | mirror sgRNA-2 | This paper; IDT | CRISPR-cas9 guide RNA | 5’-AGAAACAGGGTCGATGATGA |
| Sequence-based reagent | Genotyping Primer Forward | This paper; IDT | Genotyping primer | 5’-CGCTTGTGCCCACCTTAAAC |
| Sequence-based reagent | Genotyping Primer Reverse | This paper; IDT | Genotyping primer | 5’-GTATGGCTCGGGGGATTCTG |
| Commercial assay or kit | HCR v3 Amplifiers, Buffers | Molecular Instruments | ||
| Commercial assay or kit | DNA extraction | Omega Bio-tek | E.Z.N.A. Tissue DNA Kit | |
| Commercial assay or kit | PCR purification | Omega Bio-tek | MicroElute Cycle-Pure Kit | |
| Software, algorithm | Inference of CRISPR Edits | Synthego | RRID:SCR_0024508 |
Additional files
-
Supplementary file 1
Identification of mirror ortholog in J. coenia.
A maximum likelihood phylogeny of J. coenia, H. erato lativitta, T. castaneum, A. mellifera, and D. melanogaster Iroquois Complex genes confirms that JC_02269-RA is the ortholog of mirror.
- https://cdn.elifesciences.org/articles/96904/elife-96904-supp1-v1.docx
-
Supplementary file 2
mirror CRISPR-Cas9 injection results.
- https://cdn.elifesciences.org/articles/96904/elife-96904-supp2-v1.docx
-
Supplementary file 3
Sanger sequencing confirms mutations at CRISPR single-guide RNA (sgRNA) site in mirror 393 mosaic knockouts (mKOs).
Sanger sequencing results from three different mirror mutant individuals. Each line denotes a 394 different wild-type (WT) or mutant allele with indels indicated in red and the PAM site in green.
- https://cdn.elifesciences.org/articles/96904/elife-96904-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96904/elife-96904-mdarchecklist1-v1.pdf





