Saccharomyces cerevisiae Rev7 promotes non-homologous end-joining by blocking Mre11 nuclease and Rad50’s ATPase activities and homologous recombination
Figures

Y2H screens suggest interaction between ScRev7 and the MRX subunits.
The Y2H assay was performed in (A) wild-type, (B) rev3Δ, and (C) mre11Δ rad50Δ xrs2Δ mutant strains in PJ69-4A background. These strains were co-transformed with pairwise combinations of empty vector, bait (pGBKT7-REV7) and prey (pGADT7/MRE11, RAD50, XRS2, REV7, or SAE2) plasmids. Equal number of mid-log phase cells was spotted onto the SC/-Trp-Leu agar plates (upper panels) or SC/-Trp -Leu-His agar plates containing 3-aminotriazole (3-AT) (bottom panels). Cells were imaged after 48 hr of growth at 30°C. The images shown in panels (A–C) are representative of three independent experiments. (D) Quantitative parameters for interaction between ScRev7 and Rev1, Mre11, Rad50, Xrs2, or Mre11–Rad50 proteins.
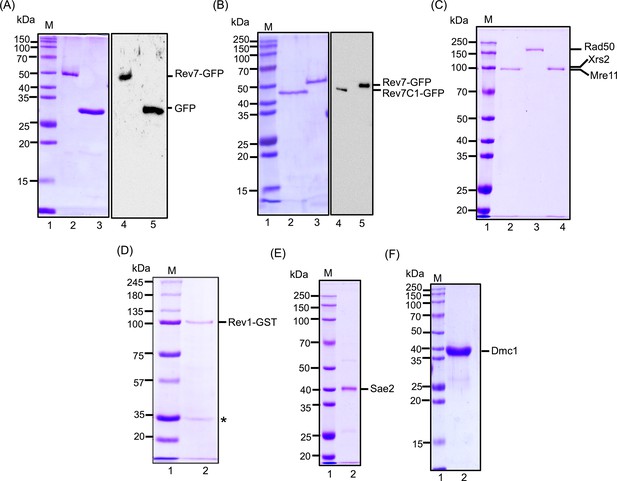
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) analysis of purified proteins used in this study.
(A) SDS–PAGE and western blot analysis of purified GFP-tagged ScRev7 and eGFP. Lane 1 (in panels A–F) standard molecular weight markers; lane 2, purified GFP-tagged ScRev7; lane 3, purified GFP. Western blot analysis of GFP-tagged Rev7 (lane 4) and eGFP (lane 5) using mouse anti-GFP antibodies. (B) Purified GFP-tagged Rev7-C1 (lane 2) and GFP-tagged Rev7 (lane 3). Western blot analysis of GFP-tagged Rev7-C1 (lane 4) and GFP-tagged Rev7 (lane 5) using mouse anti-GFP antibodies. (C) Purified MRX subunits: lane 2, Mre11; lane 3, Rad50; lane 4, Xrs2. (D) Purified GST-tagged Rev1; the faster migrating band (*) corresponds to ScRev7, which co-purifies with GST-tagged Rev1 (Acharya et al., 2006). (E) Purified Sae2 and (F) purified Dmc1.
-
Figure 1—figure supplement 1—source data 1
Original files for gel images and blots displayed in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
PDF file containing labelled uncropped gel images and blots displayed in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig1-figsupp1-data2-v1.zip
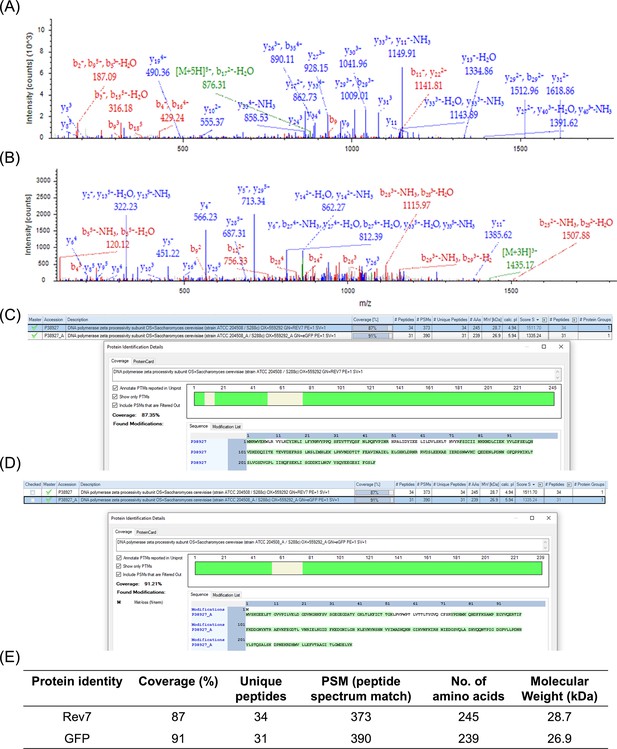
Mass spectromtery analysis of purified Rev7-GFP.
Representative spectra for unique peptides obtained after trypsin digestion of Rev7-GFP protein that matches with, (A) GFP sequence and (B) ScRev7 sequence. (C) Snapshot of sequence coverage (shown in green) obtained for Rev7 and (D) GFP. (E) Table summarizes data obtained for Orbitrap Liquid chromatography mass-spectrometry performed using purified Rev7-GFP protein.
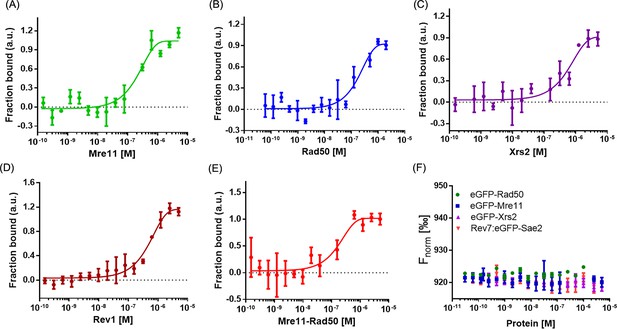
Microscale thermophoresis (MST) reveals a direct interaction between Rev7 and MRX subunits.
Normalized MST-binding curves were generated for the titration of (A) Mre11, (B) Rad50, (C) Xrs2, (D) Rev1, (E) the Mre11–Rad50 complex, and (F) Sae2 proteins against Rev7-eGFP. Panel F also includes datasets for the binding of Mre11, Rad50, and Xrs2 against eGFP. Error bars represent the standard error of the mean (SEM), and data are representative of three independent experiments. Panel F represents data from two independent experiments.
-
Figure 1—figure supplement 3—source data 1
Raw data for panels A–F.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig1-figsupp3-data1-v1.xlsx
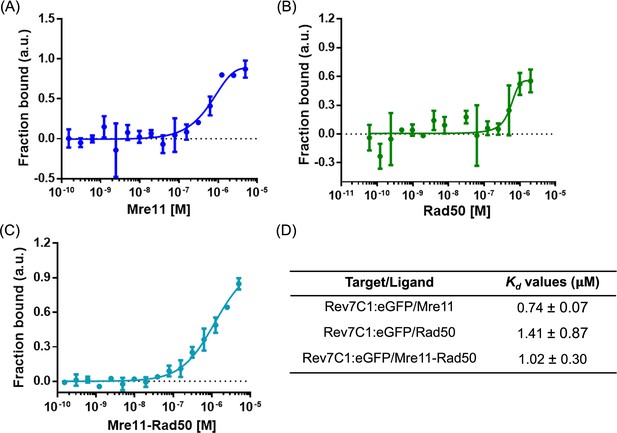
Rev7-C1 exhibits weak interactions with MRX subunits.
Binding isotherms were obtained by incubating increasing concentrations of (A) Mre11 (0.00015–5 μM), (B) Rad50 (0.00006–2 μM), and (C) MR complex (0.00015–5 μM) with fixed concentration of Rev7C1:GFP protein (0.25 μM). (D) Kd values obtained upon fitting the curves in Kd model using MO affinity analysis software (Nanotemper). Data are representative of three independent experiments.
-
Figure 1—figure supplement 4—source data 1
Raw data for panels A–C.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig1-figsupp4-data1-v1.xlsx

Deletion analysis revealed that the C-terminal 42 amino acids of ScRev7 are critical for its binding to the MRX subunits.
(A) Schematic representation of the full-length and truncated ScRev7 variants. The truncated species lacking the indicated number of amino acids (aa) in the N-terminal domain (NTDΔ) or C-terminal domain (CTDΔ) is indicated on the right-hand side of the figure. (B) Representative images of spot assays of cells carrying pairwise combination of empty vector, bait and prey plasmids expressing N-terminally truncated species of Rev7 and full-length Rev7, Mre11, Rad50, and Xrs2, respectively. (C) Same as panel (B), but with the bait plasmids encoding C-terminally truncated Rev7 variants. Cells were imaged after 48 hr of growth at 30°C. Y2H assay was performed as described in the legend to Figure 1. Data are representative of three independent experiments.
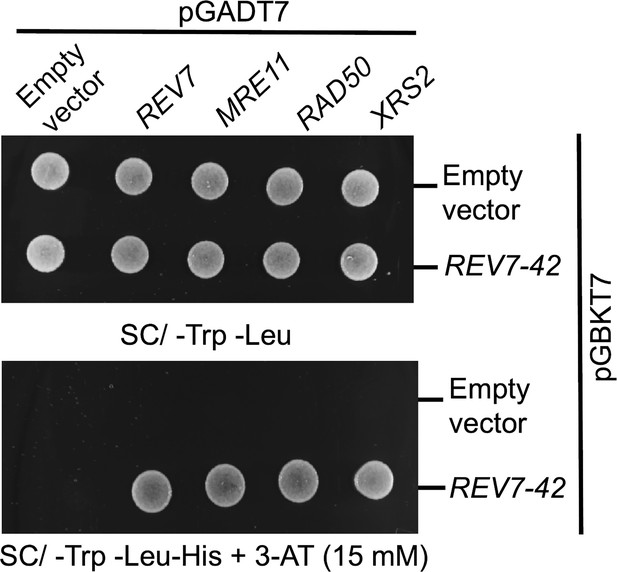
The C-terminal 42 amino-acid region of Rev7 interacts with M/R/X subunits.
The PJ69-4A cells were co-transformed with empty vectors or vectors expressing Rev7-42 residue peptide or MRX subunits as indicated. Equal number of cells was spotted onto -Trp-Leu and -Trp-Leu-His selection plates containing 15 mM 3-aminotriazole (3-AT) and incubated at 30°C for 2 days.
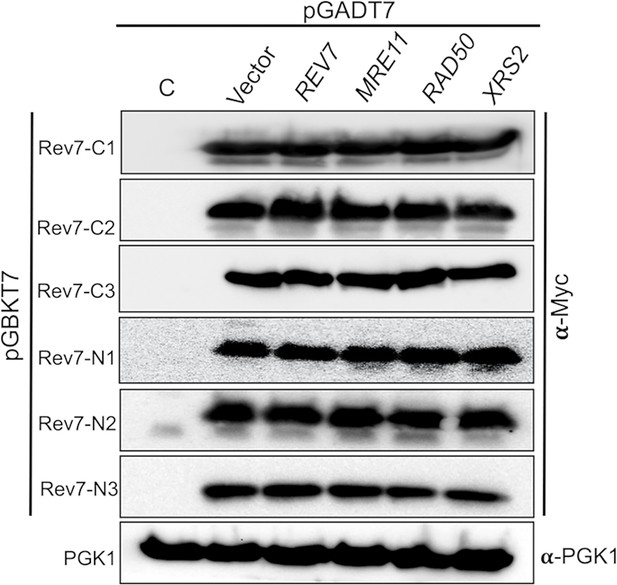
Western blot showing the abundance of N- and C-terminally truncated variants of Rev7.
S. cerevisiae PJ69-4A cells were co-transformed with the indicated bait plasmid expressing c-Myc epitope tagged N- or C-terminally truncated variant of Rev7 (Rev7-C1, Rev7-C2, Rev7-C3, Rev7-N1, Rev7-N2, or Rev7-N3) with and a prey plasmid expressing Rev7, Mre11, Rad50, or Xrs2. Pgk1 was used as the loading control and was probed with anti-Pgk1 antibodies. Sample loading was normalized to similar amounts by determining the protein concentration utilizing the dye-binding assay. Lane ‘C’ represents lysates from cells harboring empty bait and prey vectors.
-
Figure 2—figure supplement 2—source data 1
Original raw files for western blot analysis displayed in Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig2-figsupp2-data1-v1.zip
-
Figure 2—figure supplement 2—source data 2
PDF files containing uncropped labeled western blots displayed in Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig2-figsupp2-data2-v1.zip
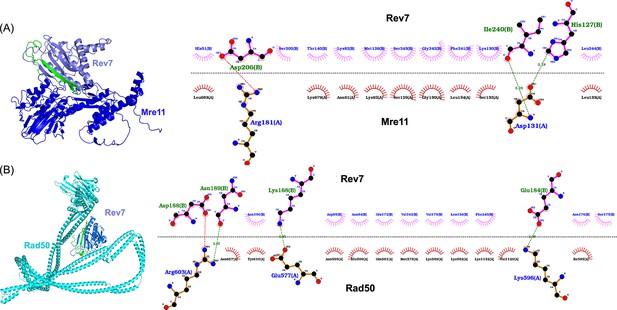
AlphaFold-Multimer generated models of Rev7–Mre11 and Rev7–Rad50 protein complexes.
(A) Model of the Rev7–Mre11 heterodimer, Rev7, light blue, Mre11, deep blue. (B) Model of Rev7–Rad50 heterodimer, Rev7, light blue, Rad50, cyan. Alongside is the two-dimensional view of the corresponding protein–protein-binding interface as generated by the LigPlot software. Numbers indicated on the dotted lines represent bond lengths in Angstroms. The C-terminal safety belt region of Rev7 is colored in green in both complexes. By convention, red, black, and blue circles represent oxygen, carbon, and nitrogen atoms, respectively. The dashed line represents the interaction interface.

A 42-amino acid segment at the extreme C-terminus of ScRev7 renders cells resistant to the synergistic adverse effect of G-quadruplex DNA and HU.
(A) REV7 deletion does not affect HU sensitivity of rev7Δ cells. Representative images of YPD plates showing spot assay of wild-type (WT) W1588-4C strain and its derivative rev7 mutant cells, in the absence or presence of 50 or 100 mM HU. (B) Representative images of SC/-Leu agar plates showing spot assay of wild-type, rev7Δ, rev7-C1, or rev7-42 aa cells harboring the indicated plasmids carrying G-quadruplex-forming motifs derived from chromosome IV (Chr.IVG4). The cells were grown on SC/-Leu agar plates in the absence of HU. (C) Same as panel (B), but the growth medium contained 100 mM HU. For serial dilutions, each strain was grown in SC/-Leu medium, normalized to OD600 = 1.0, and serially diluted using yeast nitrogen base medium. Five μl aliquots from the serial dilutions were spotted onto the SC/-Leu agar plates with or without HU. FT10/Control plasmid lacks the G-quadruplex DNA insert. The abbreviations ‘le’ and ‘lg’ stand for leading and lagging strands, respectively. Cells were imaged after 4 days of growth at 30°C. Data are representative of three independent experiments.

Purification of ScRev7 and ScRev7-C1 proteins.
(A) SDS–PAGE analysis of protein samples from different stages of ScRev7 purification. Lane 1, standard protein markers; lane 2, uninduced cell lysate (10 μg); lane 3, induced cell lysate (10 μg); lane 4, Ni2+-NTA column eluate (3 μg); lane 5, Superdex S75 column eluate (1 μg); lane 6, eluate from heparin column (0.8 μg). (B) SDS–PAGE analysis of purified truncated ScRev7-C1 variant. Lane 1: standard protein markers. Lanes 2 and 3, purified full-length ScRev7 and ScRev7-C1 variant.
-
Figure 4—source data 1
Original files for gel images displayed in Figure 4.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig4-data1-v1.zip
-
Figure 4—source data 2
PDF file containing labelled uncropped gel images displayed in Figure 4.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig4-data2-v1.zip

ScRev7 impedes both exo- and endonucleolytic activities of the MRX subunits.
(A) A representative image showing the effect of ScRev7 on the exonuclease activity of MRX complex. Reaction mixtures containing 5nM 32P-labeled 60bp dsDNA were incubated in the absence or presence of M/R/X subunits and various amounts of ScRev7. Lane 1, 32P-labeled dsDNA. Lane 2, same as in lane 1, but with 2.5 μM ScRev7. Lane 3, same as in lane 1, but with M/R/X subunits (0.1 μM each). Lanes 4–12, same as in lane 3, but with 0.05, 0.1, 0.3, 0.5, 0.7, 1, 1.5, 2, and 2.5 μM of ScRev7, respectively. (B) A representative image showing the effect of Rev7-C1 variant on the exonuclease activity of the MRX complex. Assay was performed as in panel (A), but with ScRev7-C1 variant. Lane 1, 32P-labeled dsDNA. Lane 2, same as in lane 1, but with 2.5 μM ScRev7-C1 variant. Lane 3, same as in lane 1, but with M/R/X subunits (0.1 μM each). Lanes 4 and 5, same as in lane 3, but with 1.5 and 3.0 μM of ScRev7-C1 variant, respectively. (C) A representative image showing Mre11 endonuclease activity on circular ssDNA. Reaction mixtures containing 100 ng M13 circular ssDNA were incubated without (lane 1) or with 0.1, 0.5, 1, 1.5, and 2 μM Mre11, respectively (lanes 2–6). (D) A representative image showing the effect of ScRev7 on Mre11 endonuclease activity by ScRev7. Reaction mixtures containing 1 μM Mre11 were pre-incubated with 0.5, 1.0, 1.5, and 2 μM of ScRev7 (lanes 3–6, respectively), prior to the addition of 100 ng of M13 circular ssDNA. Lane 1, circular ssDNA alone. Lane 2, same as in lane 1, but with 2 μM of ScRev7. Increasing concentrations of the indicated protein is represented by open triangles at the top of the gel.
-
Figure 5—source data 1
Original files of gels displayed in Figure 5, panels A–D.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig5-data1-v1.zip
-
Figure 5—source data 2
PDF file containing uncropped labeled gel images displayed in Figure 5.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig5-data2-v1.zip
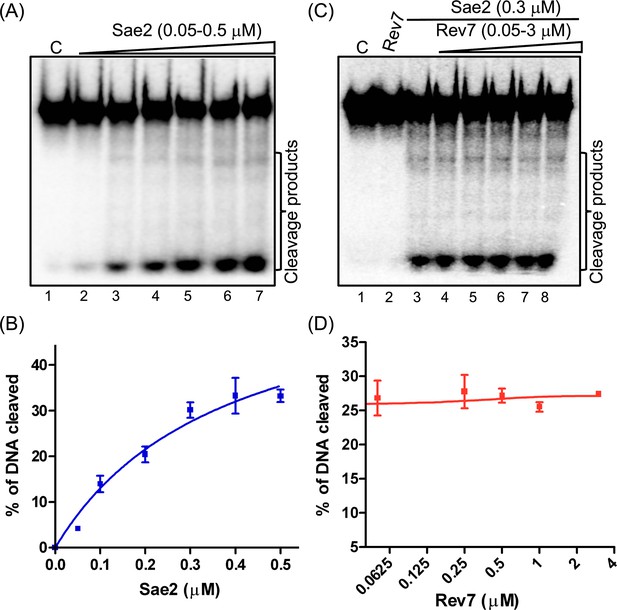
Rev7 does not impact the endonuclease activity of Sae2.
(A) Sae2-catalyzed cleavage of 32P-labeled 60 bp dsDNA. Reaction mixtures containing 5 nM dsDNA were incubated with increasing concentrations of Sae2. Lane 1: control reaction without protein; Lanes 2–7: Sae2 at 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 μM, respectively. (B) Quantification of Sae2 endonuclease activity as a function of its concentration. (C) A representative image demonstrating the effect of Rev7 on the endonuclease activity of Sae2. Lane 1: control reaction of 32P-labeled dsDNA in the absence of Sae2; lane 2: same as lane 1 but with 3 µM ScRev7; lane 3: same as lane 1 but with 0.3 µM Sae2; lanes 4–8: same as lane 3 but with increasing concentrations of ScRev7 at 0.05, 0.25, 0.5, 1.0, and 3.0 µM, respectively. (D) Quantification of the endonuclease activity catalyzed by Sae2 from panel C. The closed triangles on the top of gel images in panels (A) and (C), represent increasing concentrations of Sae2 or ScRev7, respectively. Error bars indicate SEM, and data are representative of three independent experiments.
-
Figure 5—figure supplement 1—source data 1
Original files for gels displayed in panels A and B in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
PDF file containing uncropped labeled gel images displayed in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig5-figsupp1-data2-v1.zip

Rev7 inhibits the ATPase activity of Rad50 without impacting its ability to bind ATP.
(A) Rad50 binds [γ-32P]ATP in a dose-dependent manner. Lane 1, no protein. Lanes 2–7, reactions were performed with 0.1, 0.2, 0.4, 0.6, 0.8, and 1 µM of Rad50 and 400 pmol[γ-32P]ATP. (B) ScRev7 does not affect the ability of Rad50 to bind ATP. Lane 1, Rad50 and 400 pmol[γ-32P]ATP. Lanes 2–7, same as in lane 1, but with 0.5, 1, 2, 4, 5, and 6 µM of ScRev7, respectively. (C) Quantification of ATP binding by Rad50 as a function of its concentration. (D) Quantification of the effect of ScRev7 on ATP binding by Rad50. (E) ATPase activity of Rad50 as a function of its concentration. Reactions were performed in the absence (lane 1) or presence (lanes 2–9) of 0.05, 0.1, 0.2, 0.3, 0.5, 0.6, 0.8, and 1 µM of Rad50, respectively. (F) Quantification of Rad50 ATPase activity as a function of its concentration. (G) ScRev7 abrogates ATP hydrolysis catalyzed by Rad50. (H) ScRev7 does not impact the ability of ScDmc1 to hydrolyze ATP. Lane 1 contained 400 pmol [γ-32P]ATP; lane 2, same as in lane 1, but with 2 µM of ScRev7; lane 3, as in lane 1, but with 0.5 µM of ScDmc1; lanes 4–8, as in lane 3, but with 0.3, 0.5, 1, 1.5, and 2 µM ScRev7, respectively. (I) ScRev7-C1 variant impedes ATP hydrolysis catalyzed by Rad50. In panels (G) and (I), lane 1 contained 400 pmol [γ-32P]ATP; lane 2, same as in lane 1, but 2 µM ScRev7/ScRev7-C1 variant; lane 3, as in lane 1, but with 0.5 µM Rad50; lanes 4–9, as in lane 3, but with 0.1, 0.3, 0.5, 1, 1.5, and 2 µM ScRev7/ScRev7-C1 variant, respectively. (J) Quantification of the inhibitory effect of ScRev7/ScRev7-C1 on ATP hydrolysis catalyzed by Rad50 or ScDmc1. The closed triangles on the top of gel images in panels (A), (B), (E), (G), (H), and (I) represent increasing concentrations of Rad50, ScRev7, or ScRev7-C1. Error bars indicate SEM, and data are representative of three independent experiments.
-
Figure 6—source data 1
Original files of gel images displayed in Figure 6, panels A and B.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig6-data1-v1.zip
-
Figure 6—source data 2
PDF file representing labeled uncropped gel images corresponding to Figure 6, panels A and B.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig6-data2-v1.zip
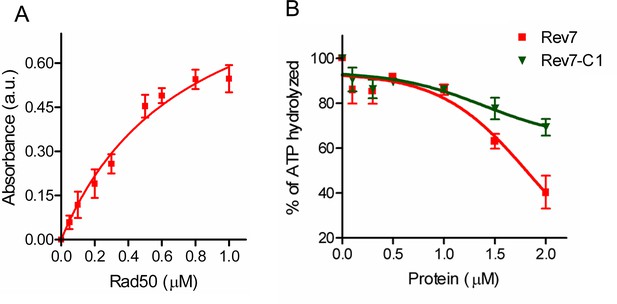
ScRev7 inhibits the ATPase activity of ScRad50.
ATPase activity was measured using malachite green phosphate detection assay that quantifies the amount of Pi released upon ATP hydrolysis. (A) Increasing concentrations of ScRad50 (0, 0.05, 0.1, 0.2, 0.3, 0.5, 0.6, 0.8, and 1 µM) were incubated with reaction mixtures containing 20 mM Tris–HCl, pH 7.5, 50 mM KCl, 5% glycerol, 0.1 mM DTT, 0.2 mg/ml Bovine Serum Albumin protein (BSA), 150 µM ATP and 1 mM MgCl2. Absorbance values were then monitored spectrophotometrically at 620 nm. (B) Same as (A), but fixed concentrations of ScRad50 (0.25 µM) were pre-incubated with increasing concentrations of ScRev7 or ScRev7-C1 (0, 0.1, 0.3, 0.5, 1, 1.5, and 2 µM). The graphs were generated by non-linear regression analysis using GraphPad Prism (V. 5.0). Error bars represent standard deviation of data from two independent experiments.

S. cerevisiae REV7 promotes non-homologous end-joining (NHEJ)-mediated double-strand break (DSB) repair.
(A) Map of the NHEJ reporter plasmid and the experimental workflow. Sixty ng of uncut or linearized pRS416 plasmid DNA was transformed into the wild-type (WT) (W1588-4C) and indicated isogenic mutants carrying single or double deletions. Transformants were selected on SC medium lacking uracil. (B) Quantification of NHEJ efficiency relative to WT cells. (C) Quantification of fold decrease in the efficiency of NHEJ relative to the WT. The boxes represent mean; whiskers, minimum and maximum values. ‘LOD’ denotes below the level of detection compared with the WT. The data are presented as the mean ± SEM of four independent experiments. n.s.: not significant, *p < 0.05, **p < 0.005, ****p < 0.0001 versus control. The exact p-values are presented in Supplementary file 1c.
-
Figure 7—source data 1
Raw data corresponding to Figure 7, panel B.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig7-data1-v1.csv
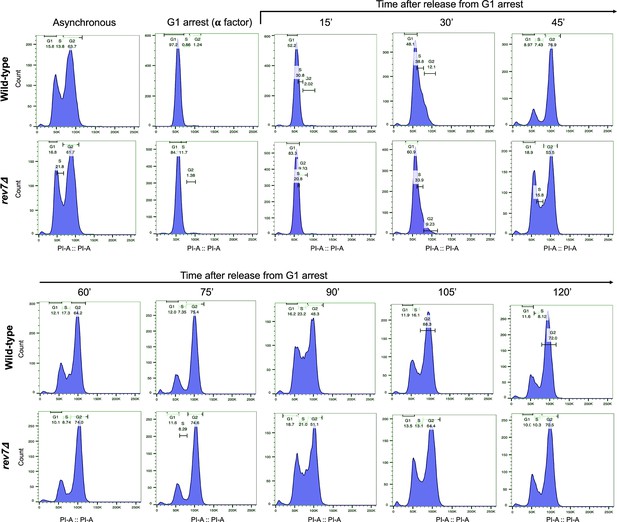
Cell cycle analysis of wild-type and rev7Δ cells.
Cell cycle progression in wild-type and rev7Δ cells was analyzed by synchronizing an equal number of cells at the G1 phase using α-factor (100 ng/ml) in the growth medium. After releasing cells from G1 arrest, they were collected at specified time points and stained with propidium iodide for fluorescence-activated cell sorting (FACS) analysis. The histograms illustrate the distribution of cells in the G1, S, and G2 phases of the cell cycle for both wild-type and rev7Δ strains. Results shown are representative of two independent experiments.

S. cerevisiae REV7 facilitates chromosomal double-strand break (DSB) repair via non-homologous end-joining (NHEJ) pathway.
(A) A schematic diagram showing the ‘suicide deletion’ cassette at the ADE2 locus of chromosome XV redrawn from Karathanasis and Wilson, 2002. The arrows indicate the locations of PCR primers. Brown and staggered red boxes correspond to direct repeats and I-SceI cleavage sites, respectively. (B) Quantification of NHEJ efficiency in the wild-type YW714 and derivative mutant strains. (C) Representative gel images of PCR-amplified DNA products from Ade2+ transformant cells. Data are means ± SEM from three independent experiments. n.s.: not significant, **p < 0.005, ***p < 0.001, ****p < 0.0001 versus control, as assessed by one-way ANOVA Dunnett’s multiple comparison test. The exact p-values are presented in Supplementary file 1d.
-
Figure 8—source data 1
Raw data corresponding to Figure 8, panel B.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig8-data1-v1.csv
-
Figure 8—source data 2
Original file and labelled PDF file for gel images displayed in Figure 8, panel C.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig8-data2-v1.zip

Deletion of REV7 increases the frequency of mitotic homologous recombination (HR).
(A) Schematic representation of plasmid-chromosome recombination assay. The ura3-1 and ura3-G4 alleles are located on chromosome V and plasmid pFAT10-G4, respectively. Recombination between a plasmid borne ura3-G4 allele and the chromosomal borne ura3-1 allele would result in Ura+ prototrophs. (B) Representative images of Ura3+ papillae on SC/-Ura agar plates in the absence or presence of 100 mM HU. (C) Quantification of the rate of HR frequency in different strains. Data are presented as mean ± SD from three different experiments. ns, not significant, **p < 0.01, ***p < 0.001, ****p < 0.0001 versus control, as assessed using one-way ANOVA and Tukey’s post hoc test.
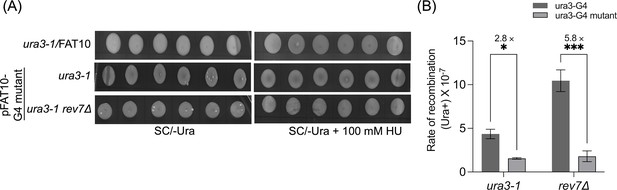
Mutation of G-quadruplex-forming motifs markedly attenuate the rate of homologous recombination (HR) frequency in rev7Δcells.
(A) Representative images showing Ura3+ papillae on SC/-Ura agar plates in the absence or presence of 100 mM HU. Cells were imaged after 6 days of growth at 30°C. (B) Quantification of the rate of HR frequency in ura3-1 and ura3-1 rev7Δ strains carrying plasmids with unmutated and mutated ura3-G4 inserts in the absence of HU. Data are presented as mean ± SD from three different experiments. n.s., not significant, *p < 0.05, and ***p < 0.001 versus control, as assessed using two-way ANOVA and Sidaks multiple comparison test.
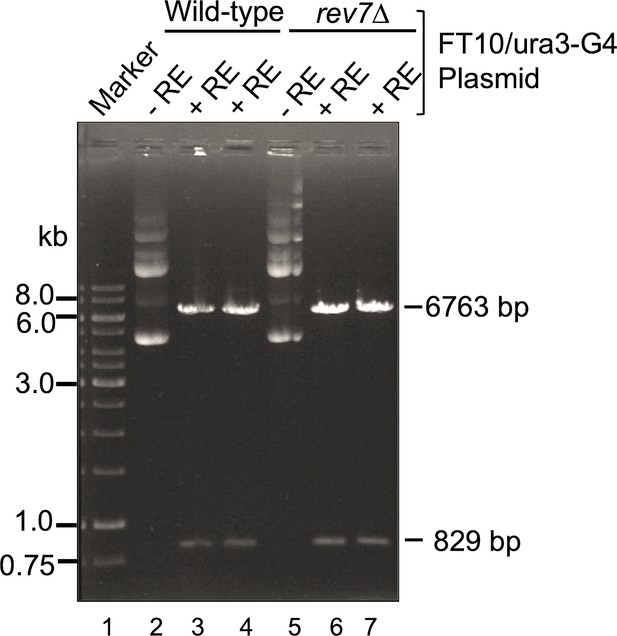
G-quadruplex-forming motifs are stable in rev7Δ cells during the homologous recombination (HR) assay.
Total plasmid pFAT10-G4 DNA was isolated from cultures of wild-type (WT) and rev7∆ cells. Samples of undigested and BamHI and Sph1 digested plasmid pFAT10-G4 DNA were analyzed by electrophoresis on a 0.6% agarose gel and visualized after staining with ethidium bromide. Lane1: DNA marker; lanes 2 and 5, represent undigested plasmid DNA from the WT and rev7∆ cells, respectively. Lanes 3 and 4, plasmid pFAT10-G4 DNA from the WT cells digested by BamHI and Sph1; lanes 6 and 7, same as lanes 3 and 5, but plasmid isolated from rev7∆ cells. +RE and −RE indicate DNA digested by BamHI and Sph1 and undigested plasmid DNA, respectively.
-
Figure 9—figure supplement 2—source data 1
Original file and labeled PDF file for the gel image displayed in Figure 9—figure supplement 2.
- https://cdn.elifesciences.org/articles/96933/elife-96933-fig9-figsupp2-data1-v1.zip
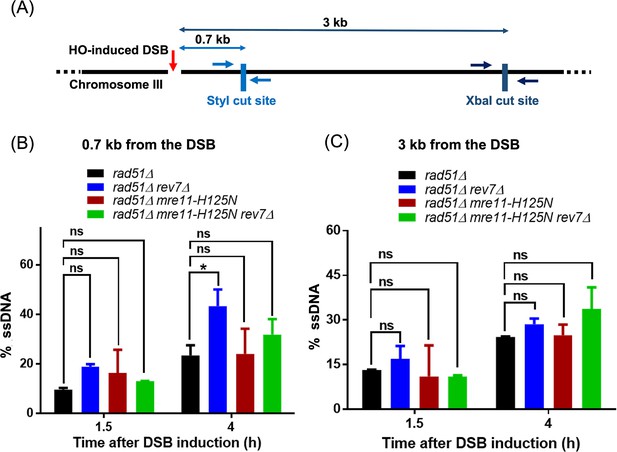
Deletion of Rev7 enhances the speed of short-range end-resection in S. cerevisiae.
(A) Schematic diagram represents the positions of StyI (0.7 kb) and XbaI (3 kb) restriction sites with respect to HO endonuclease-induced double-strand break (DSB) at the Chromosome III MAT locus. Arrows flanking the restriction sites mark the binding sites of forward and reverse primers used for qPCR analysis. The 5'—3' end-resection was physically assessed in the indicated strains by quantifying the percentage of ssDNA generated at 1.5 and 4 hr post induction of DSB, at a distance of (B) 0.7 kb and (C) 3 kb from the DSB. Two-way ANOVA was performed to determine the statistical significance of the datasets and graphs were generated using GraphPad Prism (Version 5.0). Data are representative of two independent biological replicates. ns, not significant; * p < 0.05.
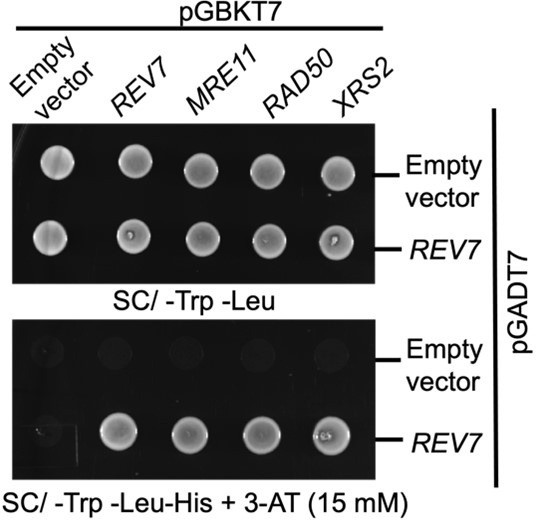
Yeast two hybrid analysis suggest interaction between Rev7 and MRX subunits.
PJ69-4A cells were co-transformed with bait vector expressing Rev7 or the Mre11, Rad50 or Xrs2 subunits and prey vector expressing Rev7 protein. Equal number of cells were spotted onto –Trp – Leu and –Trp – Leu –His dropout plates containing 3-AT and images were obtained following 48 h of incubation at 30°C. The data is representative of three independent experiments.
Additional files
-
Supplementary file 1
Inter-atomic distances and confidence parameters of amino acid residues mediating Rev7–Mre11 interactions.
Residues in bold are present in the C-terminal safety-belt region of Rev7 protein.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp1-v1.docx
-
Supplementary file 2
Inter-atomic distances and confidence parameters of amino acid residues mediating Rev7–Rad50 interactions.
Residues in bold are present in the C-terminal safety-belt region of Rev7 protein.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp2-v1.docx
-
Supplementary file 3
The exact p-values for Figure 7B.
The p-values were obtained by comparing the percentage of non-homologous end-joining observed for the indicated single- or double-gene deletions versus either the wild-type (WT) or rev7Δ strain, using non-parametric one-way ANOVA Dunnett test.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp3-v1.docx
-
Supplementary file 4
The exact p-values for Figure 8B.
The p-values were obtained by comparing the percentage of non-homologous end-joining observed for the indicated single- or double-gene deletions versus either the wild-type (WT) or rev7Δ strain, using non-parametric one-way ANOVA Dunnett test.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp4-v1.docx
-
Supplementary file 5
S. cerevisiae strains used in this study.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp5-v1.docx
-
Supplementary file 6
Sequences of primers used in this study.
The bold letters correspond to restrictions sites.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp6-v1.docx
-
Supplementary file 7
Sequences of oligonucleotides used in the preparation of DNA substrates.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp7-v1.docx
-
Supplementary file 8
Primers used for qPCR analysis.
- https://cdn.elifesciences.org/articles/96933/elife-96933-supp8-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96933/elife-96933-mdarchecklist1-v1.pdf





