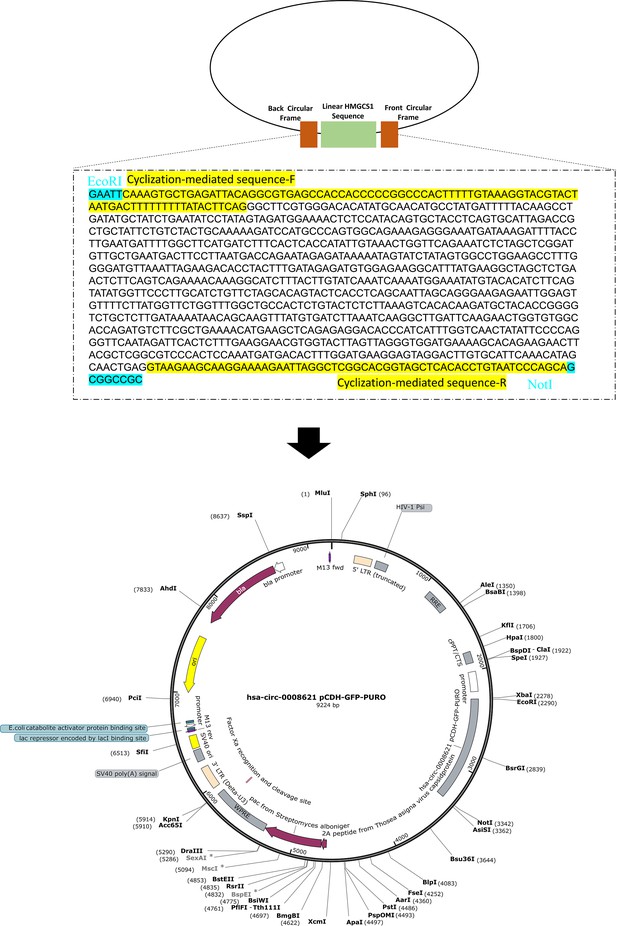
Circular RNA HMGCS1 sponges MIR4521 to aggravate type 2 diabetes-induced vascular endothelial dysfunction
Figures
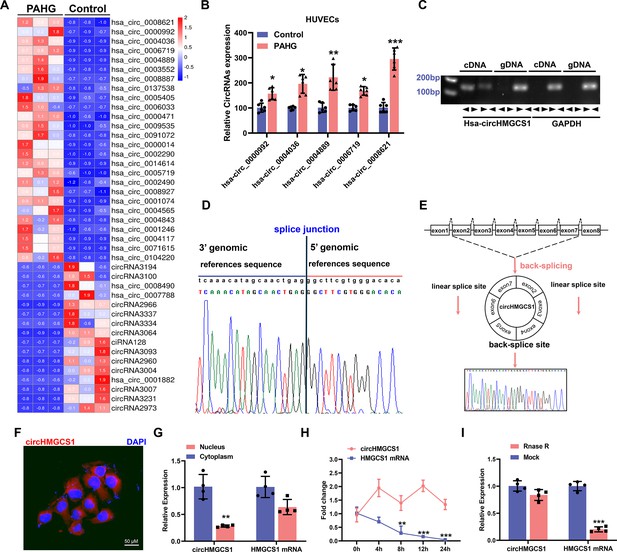
circHMGCS1 upregulation and its association with PAHG-induced endothelial dysfunction.
(A) Heat map illustrating the differential expression of circRNAs in HUVECs treated with PAHG for 24 hours (n=3). (B) qRT-PCR validation of five circRNAs in HUVECs treated with PAHG compared to normal cells, normalized to GAPDH (n=6). (C) PCR validation of circHMGCS1 presence in HUVECs, GAPDH was utilized as a negative control (n=6). (D) Alignment of circHMGCS1 sequence in CircBase (circular RNA database, upper) in agreement with Sanger sequencing results (lower). (E) Schematic representation of circularization of exons 2–7 of HMGCS1 (red arrow). (F) RNA-FISH analysis detecting circHMGCS1 expression in HUVECs using Cy3-labeled probes, Nuclei were counterstained with DAPI (red represents circHMGCS1, blue represents nucleus, scale bar=50 μm, n=4). (G) qRT-PCR quantification of circHMGCS1 and HMGCS1 mRNA levels in the cytoplasm or nucleus of HUVECs. circHMGCS1 and HMGCS1 mRNA levels were normalized to cytoplasmic values (n=4). (H) qRT-PCR measure circHMGCS1 and HMGCS1 mRNA levels after actinomycin D treatment (n=4 at different time points). (I) circHMGCS1 expression was detected in RNase R-treated total RNA by qRT-PCR (n=4). *p<0.05, **p<0.01, ***p<0.001, All significant difference was determined by unpaired two-tailed Student’s t-test or one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 1—source data 1
Uncropped and labeled gels for (Figure 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig1-data1-v1.zip
-
Figure 1—source data 2
Raw unedited gels for (Figure 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig1-data2-v1.zip
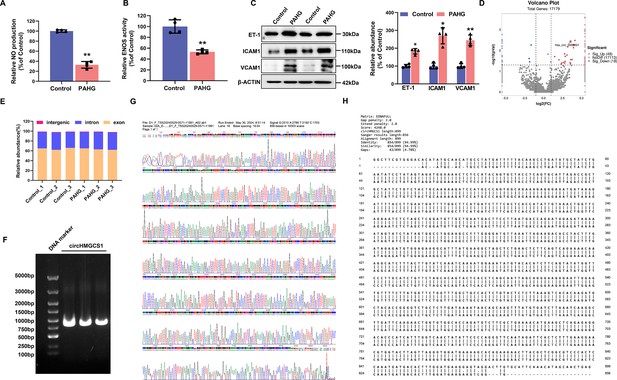
Distinct expression profiles of circRNAs in endothelial dysfunction of HUVECs.
HUVECs exposed to palmitic acid and high glucose (PAHG) treatment for 24 hr were compared with untreated Control. (A) Alterations in NO content between the control and PAHG-treated groups (n=4). (B) Changes in ENOS activity between the control and PAHG-treated groups (n=4). (C) Relative expression levels of adhesion molecules (ICAM1, VCAM1, and ET-1) in HUVECs were assessed by western blotting between the control and PAHG-treated groups (n=4). (D) Volcano plot illustrating significantly altered circRNAs between the control and PAHG-treated groups. Fold-change>2 or<0.5, p≤0.001. (E) Number of identified circRNAs in Control groups and PAHG-treated groups (n=3). (F) PCR amplification confirming the full-length circHMGCS1 (n=3). (G) Sanger sequencing validation of the full-length circHMGCS1 sequence (n=3). (H) Comparison results of circHMGCS1 sequences in Sanger sequencing and circbase database. *p<0.05, **p<0.01, All significant difference was determined by unpaired two-tailed Student’s t-test, error bar indicates SD.
-
Figure 1—figure supplement 1—source data 1
Uncropped and labeled gels for (Figure 1—figure supplement 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Raw unedited gels for (Figure 1—figure supplement 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig1-figsupp1-data2-v1.zip
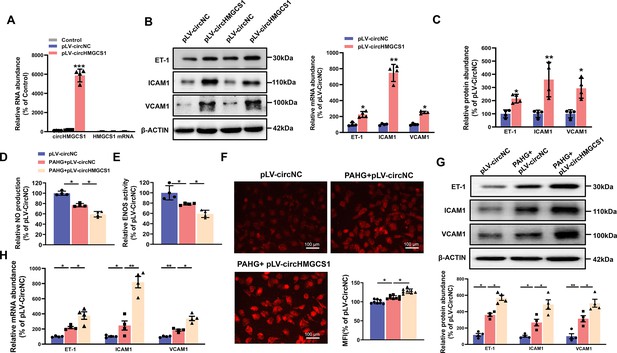
circHMGCS1 overexpression aggravates PAHG-induced endothelial dysfunction.
(A) HUVECs were transfected with either circHMGCS1 overexpression lentivirus (pLV-circHMGCS1) or lentiviral circular RNA negative control vector (pLV-circNC). After 48 hours of infection, circHMGCS1 and HMGCS1 expression levels were assessed by qRT-PCR and normalized to GAPDH (n=4). (B, C) Western blot and qRT-PCR were conducted to detect the expressions of adhesion molecules (VCAM1, ICAM1, and ET-1) in HUVECs between pLV-circHMGCS1 and pLV-circNC groups (n=4). (D) NO content in pLV-circHMGCS1-infected HUVECs after PAHG treatment (n=4). (E) ENOS activity in pLV-circHMGCS1-infused HUVECs after PAHG treatment (n=4). (F) DHE fluorescence was used to characterize ROS expression in different groups. Scale bar=100 μm (n=8). (G, H) Relative expression of adhesion molecules (ICAM1, VCAM1 and ET-1) in HUVECs infected with pLV-circNC or pLV-circHMGCS1 after PAHG treatment were determined by Western blot and qRT-PCR (n=4). *p<0.05, **p0.01, ***p<0.001, All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 2—source data 1
Uncropped and labeled gels for (Figure 2).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig2-data1-v1.zip
-
Figure 2—source data 2
Raw unedited gels for (Figure 2).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig2-data2-v1.zip
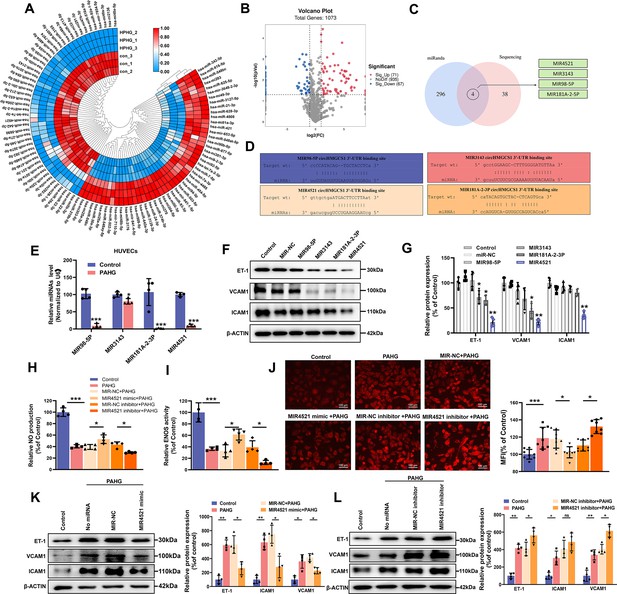
MIR4521 prevents PAHG-induced endothelial dysfunction.
(A) Heat map depicting differentially expressed miRNAs in HUVECs with or without PAHG treatment for 24 hr (n=3). (B) Volcano plot illustrating significant changes in miRNAs between control and PAHG-treated groups (Fold-change>2 or<0.5, p≤0.01). (C) Schematic illustration showing overlapping target miRNAs of circHMGCS1 predicated by miRanda and sequencing results. (D) The binding sites of circHMGCS1 were predicted using miRanda and involve MIR98-5P, MIR3143, MIR4521, and MIR181A-2–3 P. (E) Relative expression of four miRNA candidates in HUVECs treated with PAHG was assessed using qRT-PCR (n=4). (F, G) The regulatory effects of four miRNA mimics on the protein expression levels of adhesion molecules (ICAM1, VCAM1, and ET-1) (n=4). (H) Regulation of NO content by MIR4521 mimic or MIR4521 inhibitor under PAHG treatment (n=4). (I) Impact of MIR4521 mimic or MIR4521 inhibitor on ENOS activity during PAHG treatment. (n=4). (J) The ROS expression in MIR4521 mimic or MIR4521 inhibitor combined with PAHG treatment (Scale bar=100 µm, n=8). (K) Adhesion molecules (ICAM1, VCAM1, and ET-1) expression in PAHG-treated HUVECs transfected with MIR4521 mimics was determined by Western blot (n=4). (L) Determination of adhesion molecules (ICAM1, VCAM1, and ET-1) expression via Western blot in PAHG-treated HUVECs transfected with MIR4521 inhibitor (n=4). *p<0.05, **p<0.01, ***p<0.001, All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 3—source data 1
Uncropped and labeled gels for (Figure 3).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig3-data1-v1.zip
-
Figure 3—source data 2
Raw unedited gels for (Figure 3).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig3-data2-v1.zip
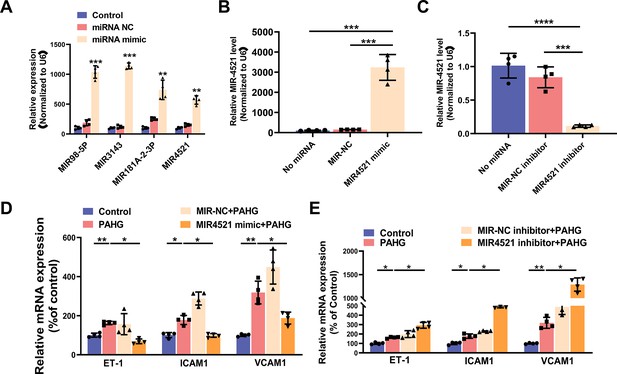
MIR4521 inhibition aggravates diabetes-induced endothelial dysfunction.
(A) Relative expression of MIR98-5P, MIR3143, MIR181A-2–3 P and MIR4521 were detected in HUVECs after corresponding miRNA mimics treatment by qRT-PCR (n=4). (B, C) Relative expression level of MIR4521 was determined by qRT-PCR in HUVECs transfected with MIR4521 mimics or MIR4521 inhibitor (n=4). (D) MIR4521 mimic inhibited the expression of adhesion molecules (ICAM1, VCAM1, and ET-1) in PAHG-treated HUVEC as detected by qRT-PCR (n=4). (E) MIR4521 inhibitor exacerbated the expression of adhesion molecules (ICAM1, VCAM1, and ET-1) in PAHG-treated HUVEC as detected by qRT-PCR (n=4). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
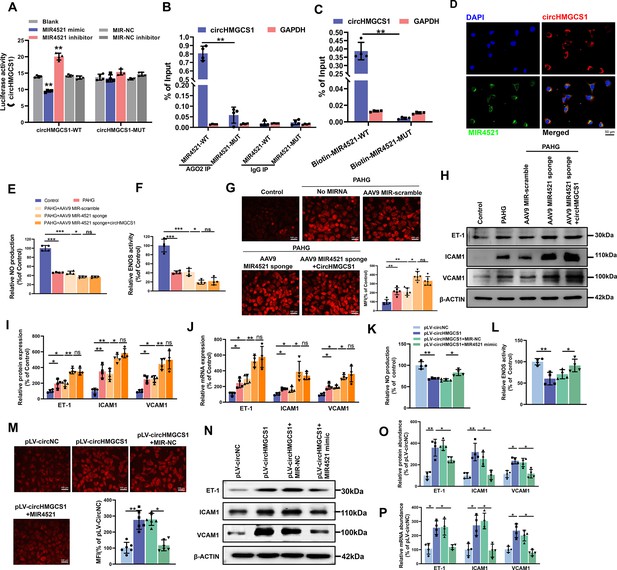
circHMGCS1 regulates PAHG-induced endothelial dysfunction by targeting and sponging MIR4521.
(A) Luciferase reporter constructs containing wild-type (WT) or mutant (MUT) circHMGCS1 were cotransfected with MIR4521 mimics, MIR-NC, MIR4521 inhibitor, or MIR-NC inhibitor in HEK293T cells (n=4). (B) Immunoprecipitation shows AGO2-mediated binding of circHMGCS1 and MIR4521 (n=4). (C) Biotin-coupled MIR4521 or its mutant probe was employed for circHMGCS1 pull-down, and captured circHMGCS1 level was quantified by qRT-PCR (n=4). (D) RNA-FISH showing the colocalization of circHMGCS1 and MIR4521 in HUVECs (red represents circHMGCS1, green represents MIR4521, blue represents nucleus, scale bar=50 μm, n=4). (E, F) circHMGCS1 exhibited no impact on NO content and ENOS activity in the presence of MIR4521 sponge (n=4). (G) circHMGCS1 demonstrated no influence on ROS expression with MIR4521 sponge (Scale bar=100 µm, n=8). (H–J) circHMGCS1 had no effect on the expression of adhesion molecules (ICAM1, VCAM1, and ET-1) in the presence of MIR4521 sponge which were determined by western blot and qRT-PCR (n=4). (K, L) MIR4521 attenuated the reduction of NO content and ENOS activity induced by circHMGCS1 (n=4). (M) MIR4521 inhibited the increase of ROS expression caused by circHMGCS1 (Scale bar=100 µm, n=8). (N–P) MIR4521 attenuated the increased expression of adhesion molecules (ICAM1, VCAM1, and ET-1) induced by circHMGCS1 which were determined by Western blot and qRT-PCR, respectively (n=4). *p<0.05, **p<0.01, ns means no significant. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 4—source data 1
Uncropped and labeled gels for (Figure 4).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig4-data1-v1.zip
-
Figure 4—source data 2
Raw unedited gels for (Figure 4).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig4-data2-v1.zip
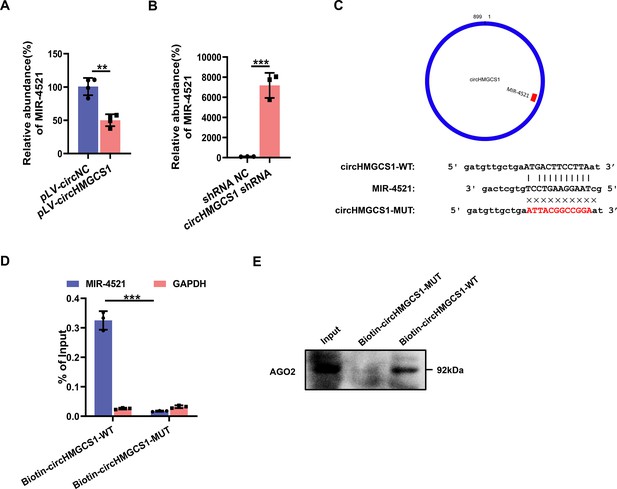
Verification of the binding site of MIR4521 and circHMGCS1.
HEK293T cells were used to package lentivirus overexpressing circHMGCS1, and then infected HUVECs cells, (A) Expression level of MIR4521 was assessed by qRT-PCR in pLV-circHMGCS1-transfected HEK293T cells (n=4). (B) The expression level of MIR4521 was assessed by qRT-PCR in HUVEC cells transfected with circHMGCS1 shRNA (n=3). (C) Schematic illustration of the predicted binding sites of MIR4521 on the circHMGCS1 transcript in human cells. (D) Biotin-coupled circHMGCS1 or its mutant probe was employed for MIR4521 pull-down assay, and captured MIR4521 level was quantified by qRT-PCR (n=4). (E) Western blot analysis of AGO2 protein expression captured by biotin-coupled circHMGCS1. **p<0.01, ***p<0.001, significant difference was determined by unpaired two-tailed Student’s t-test, error bar indicates SD.
-
Figure 4—figure supplement 1—source data 1
Uncropped and labeled gels for (Figure 4—figure supplement 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Raw unedited gels for (Figure 4—figure supplement 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig4-figsupp1-data2-v1.zip
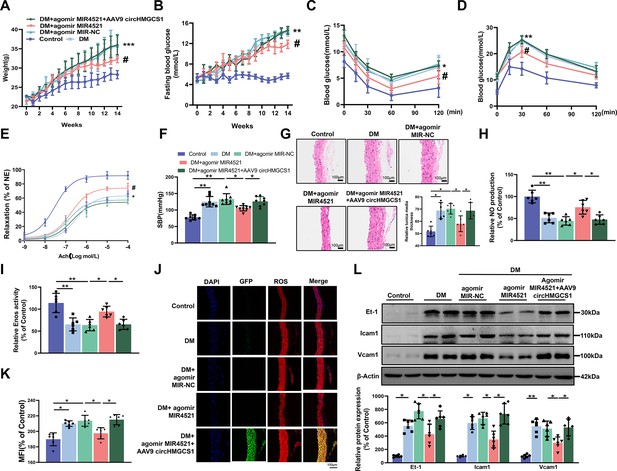
AAV9-mediated circHMGCS1 overexpression attenuates the protective effect of MIR4521 against diabetes-induced VED.
(A) Changes in body weight of mice from 0 to 14 weeks (n=8, ***p<0.001 DM versus control, #p<0.05 DM+agomir MIR4521 versus DM). (B) Alterations in fasting blood glucose in mice from 0 to 14 weeks (n=8, ***p<0.001 DM versus control, #p<0.05 DM+agomir MIR4521 versus DM). (C, D) ITT and OGTT were performed in the fasted mice (n=8, *p<0.05 DM versus control, #p<0.05 DM+agomir MIR4521 versus DM). (E) Relaxation responses of aortic rings from control, MIR4521 agomir, or MIR4521 agomir +AAV9 circHMGCS1 mice on DM diet (n=6). (F) SBP measured via tail-cuff method in different groups (n=8). (G) Hematoxylin and eosin staining (H&E) was performed on serial cross-sections of thoracic aortas from differently treated mice to assess vessel wall thickness. Scale bar=100 μm (n=6). (H, I) NO content and Enos activity in thoracic aorta after 6 weeks of DM diet with MIR4521 agomir or MIR4521 agomir +circHMGCS1 treatment (n=6). (J, K) Detection of ROS expression in thoracic aorta using DHE after MIR4521 agomir or MIR4521 agomir +AAV9 circHMGCS1 injection (red represents ROS, green represents GFP, blue represents nucleus, scale bar=100 μm, n=6). (L) Expression levels of adhesion molecules (Icam1, Vcam1, and Et-1) in thoracic aorta after 6 weeks of DM diet with MIR4521 agomir or MIR4521 agomir +circHMGCS1 treatment (n=6). *p<0.05, **p<0.01, ***p<0.001. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 5—source data 1
Uncropped and labeled gels for (Figure 5).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig5-data1-v1.zip
-
Figure 5—source data 2
Raw unedited gels for (Figure 5).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig5-data2-v1.zip
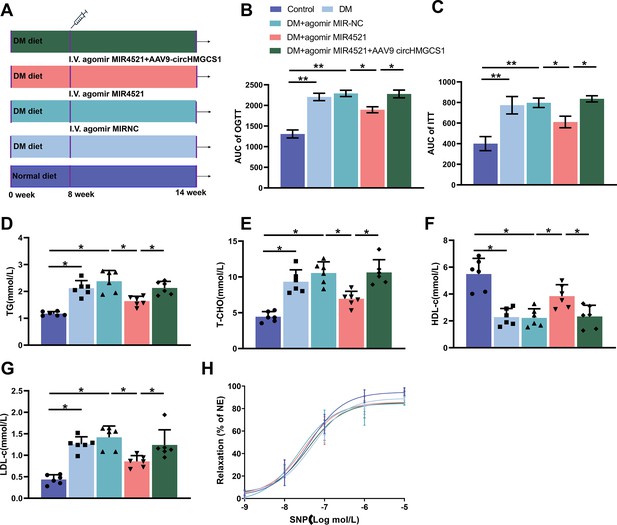
circHMGCS1 counteracts the protective role of MIR4521 in diabetes-induced VED.
(A) Timeline illustrating the HFHG feeding, AAV9 circHMGCS1, and MIR4521 agomir treatment schedule. (B, C) Quantification of the area under the curve (AUC) for glucose and insulin tests (n=8). (D) Triglyceride (TG), (E) Total cholesterol (T-CHO), (F) High-density lipoprotein cholesterol (HDL-C), and (G) Low-density lipoprotein cholesterol (LDL-C) levels in the serum were measured using Automatic Biochemical Analyzer (n=6). (H) Effects on endothelium-independent vasorelaxation to SNP (n=6). *p<0.05, **p<0.01. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
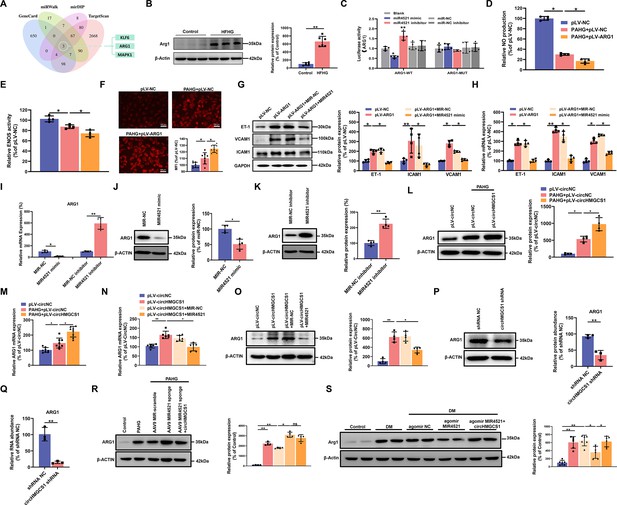
circHMGCS1 functions as a MIR4521 sponge in HUVECs to modulate ARG1 expression.
(A) Schematic depiction illustrating the intersection of predicted target genes of MIR4521 from TargetScan, miRWalk, mirDIP, and GeneCard. (B) Arg1 expression in thoracic aorta of DM by western blot (n=6). (C) HEK293T cells were cotransfected with MIR4521 mimics, MIR-NC, MIR4521 inhibitor, or MIR-NC inhibitor, along with luciferase reporter constructs containing WT or MUT 3′-untranslated region of ARG1 (n=4). (D, E) Modulatory effect of MIR4521 mimic on NO content and ENOS activity under ARG1 overexpression (n=4). (F) Regulatory impact of MIR4521 mimic on ROS content under ARG1 overexpression (Scale bar=100 µm, n=4). (G, H) ARG1-overexpressing HUVECs were generated using lentivirus and transfected with MIR4521 mimics for 24 hr to evaluate adhesion molecule expression (ICAM1, VCAM1, and ET-1) by western blot and qRT-PCR (n=4). (I–K) ARG1 expression was significantly decreased by MIR4521 mimics and increased by MIR4521 inhibitor, as determined by qRT-PCR and Western blot (n=4). (L, M) ARG1 expression was significantly increased by circHMGCS1 overexpression, as determined by western blot and qRT-PCR (n≥4). (N, O) circHMGCS1-overexpressed HUVECs, created with lentivirus and transfected with MIR4521 mimics for 48 hr, were examined for ARG1 expression by western blot and qRT-PCR (n=4). (P, Q) ARG1 expression was significantly reduced by circHMGCS1 shRNA, as determined by western blot and qRT-PCR (n=3). (R) AAV9 MIR4521 sponge was used to inhibit the MIR4521 expression in HUVECs, followed by circHMGCS1 transfection, and then treated with PAHG for 24 hr to evaluate the expression of ARG1 by western blot (n=4). (S) Detection of Arg1 expression in the thoracic aorta of DM treated with AAV9 MIR4521 agomir combined with AAV9 circHMGCS1 by western blot (n=4). *p<0.05, **p<0.01, ns means no significant. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 6—source data 1
Uncropped and labeled gels for (Figure 6).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig6-data1-v1.zip
-
Figure 6—source data 2
Raw unedited gels for (Figure 6).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig6-data2-v1.zip
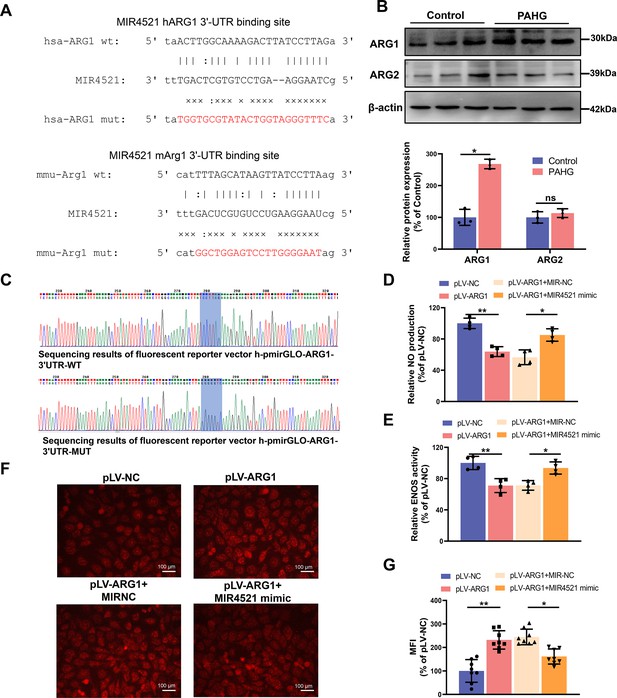
MIR4521 modulates the expression of ARG1.
(A) Schematic representation of the predicted binding sites and mutated sites for MIR4521 on ARG1 transcripts in both human cells and mice. (B) Western blot analysis assessing ARG1 and ARG2 expression in HUVECs after PAHG treatment (n=3). (C) Sequencing results displaying the mutation site in the dual luciferase assay. (D) Relative NO content. (E) Relative ENOS activity (n=4). (F, G) Expression and relative fluorescence intensity analysis of ROS detected by DHE probe (Scale bar=100 μm, n=4). *p<0.05, **p<0.01, ns means no significant. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 6—figure supplement 1—source data 1
Uncropped and labeled gels for (Figure 6—figure supplement 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Raw unedited gels for (Figure 6—figure supplement 1).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig6-figsupp1-data2-v1.zip
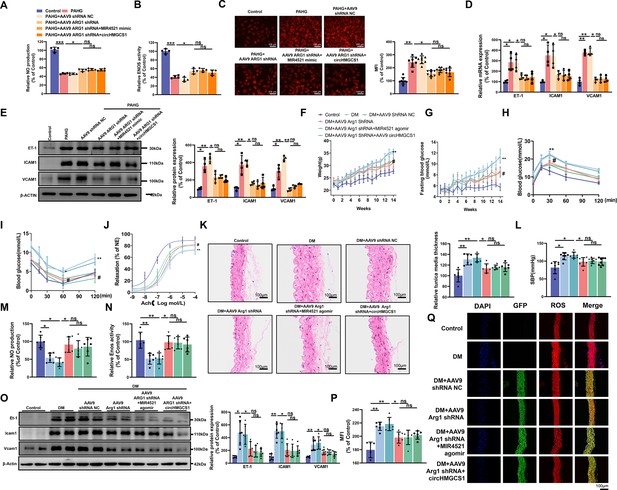
ARG1 is inseparable from circHMGCS1 and MIR4521 regulating diabetes-induced VED.
(A, B) circHMGCS1 and MIR4521 had no effect on NO content and ENOS activity expression in the absence of ARG1 (n=4). (C) ROS expression remained unaffected by circHMGCS1 and MIR4521 in the absence of ARG1 (n=4). (D, E) Relative expression of adhesion molecules (ICAM1, VCAM1, and ET-1) remained unchanged in the absence of ARG1, despite the presence of circHMGCS1 and MIR4521, as determined by qRT-PCR and Western blot (n=4). (F) Changes in mice body weight over the experimental period (n=8, **p<0.01, DM versus control, #p<0.05, DM +AAV9 ARG1 shRNA versus DM). (G) Fasting blood glucose levels in mice over time (n=8, **p<0.01, DM versus control, #p<0.05, DM +AAV9 ARG1 shRNA versus DM). (H, I) Blood glucose levels measured at week 13 (ITT) and week 14 (OGTT) (n=8, **p<0.01 DM versus control, #p<0.05, DM +AAV9 ARG1 shRNA versus DM). (J) Endothelium-dependent relaxations in aortic rings from different groups (n=8). (K) H&E performed on serial cross-sections of thoracic aortas from differently treated mice to evaluate vessel wall thickness. Scale bar=100 μm (n=6). (L) SBP measured by the tail-cuff method in different groups (n=8). (M, N) NO content and Enos activity expression in thoracic aort (n=6). (O) Relative expression of adhesion molecules (Icam1, Vcam1, and Et-1) in thoracic aorta assessed by Western blot (n=6). (P, Q) ROS expression in the thoracic aorta using the DHE probe (Red represents ROS, Green represents GFP, Blue represents nucleus, scale bar=100 μm, n=6). *p<0.05, **p<0.01, ns means no significant. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.
-
Figure 7—source data 1
Uncropped and labeled gels for (Figure 7).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig7-data1-v1.zip
-
Figure 7—source data 2
Raw unedited gels for (Figure 7).
- https://cdn.elifesciences.org/articles/97267/elife-97267-fig7-data2-v1.zip
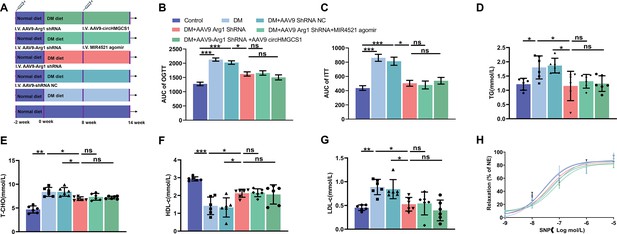
Loss of functional regulation of circHMGCS1 and MIR4521 on HUVECs in the absence of ARG1 in vivo.
(A) Schematic outlining the timeline of HFHG feeding, AAV9 circHMGCS1 and MIR4521 agomir treatment, AAV9 ARG1 shRNA treatment, and experiment conclusion. (B, C) Blood glucose levels measured at week 13 (ITT) and week 14 (OGTT), the corresponding AUC for glucose or insulin tests was calculated (n=8, **p<0.01, DM versus control, #p<0.05, DM +AAV9 ARG1 shRNA versus HFHG). (D) TG, (E) T-CHO, (F) HDL-C, and (G) LDL-C levels in the serum of different treated groups (n=6). (H) Assessment of endothelium-independent vasorelaxation to SNP (n=6). *p<0.05, **p<0.01, ***p<0.001, ns means no significant. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.

The impact of circHMGCS1 knockdown on ARG1 and miR-4521 expression levels in HUVEC.
The cells were transfected with either circHMGCS1 shRNA1 or circHMGCS1 shRNA2, and the expressions levels of circHMGCS1 and HMGCS1 (A), miR-4521 (B) and ARG1 (C and D) in HUVECs were detected by qRT-PCR and Western blot. n=3 in each group. *p < 0.05, **p < 0.01. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.

The impact of circHMGCS1 knockdown on ARG1 and miR-4521 expression levels in HUVEC.
The cells were transfected with either circHMGCS1 shRNA1 or circHMGCS1 shRNA2, and the expressions levels of circHMGCS1 and HMGCS1 (A), miR-4521 (B) and ARG1 (C and D) in HUVECs were detected by qRT-PCR and Western blot. n=3 in each group. *p < 0.05, **p < 0.01. All significant difference was determined by one-way ANOVA followed by Bonferroni multiple comparison post hoc test, error bar indicates SD.

The distribution of copy numbers for circHMGCS1, miR-4521 and ARG1 in HUVECs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HUVEC | The Shanghai Cell Bank of the Chinese Academy of Sciences | RRID:CVCL_2959 | |
| Cell line (H. sapiens) | HEK-293T | The Shanghai Cell Bank of the Chinese Academy of Sciences | RRID:CVCL_0063 | |
| Gene (H. sapiens) | circHMGCS1 | circbase | circRNA ID: hsa_circ_0008621 | |
| Gene (H. sapiens) | HMGCS1 | GeneBank | Gene ID: 3157 | |
| Gene (H. sapiens) | MIR4521 | miRbase | miRNA ID: MIMAT0019058 | |
| Gene (Mus musculus) | ARG1 | GeneBank | Gene ID: 383 | |
| Gene (M. musculus) | Arg1 | GeneBank | Gene ID: 11846 | |
| Antibody | Anti- Liver Arginase (ARG1) (Rabbit monoclonal) | Abcam | ab133543 | WB (1: 1000) |
| Antibody | Anti- Arg2 (Rabbit monoclonal) | Abcam | ab264066 | WB (1: 1000) |
| Antibody | Anti- Endothelin 1(ET-1)(Mouse monoclonal) | Abcam | ab2786 | WB (1: 1000) |
| Antibody | Anti- VCAM1 (Rabbit monoclonal) | Abcam | ab134047 | WB (1: 1000) |
| Antibody | Anti-ICAM1 (Rabbit monoclonal) | Abcam | ab222736 | WB (1: 1000) |
| Antibody | Anti-AGO2 (Mouse monoclonal) | Proteintech | 67934–1-Ig | WB (1: 2000) |
| Antibody | Anti-beta ACTIN (Mouse monoclonal) | Abcam | ab8226 | WB (1: 2000) |
| Antibody | Anti-GAPDH (Mouse monoclonal) | Abcam | ab8245 | WB (1: 2000) |
| Recombinant DNA reagent | pCDH-CMV-MCS-EF1-Puro (plasmid) | Addgene | RRID:Addgene_73030 | |
| Recombinant DNA reagent | pLKO.1-GFP-shRNA (plasmid) | Addgene | RRID:Addgene_30323 | |
| Recombinant DNA reagent | psPAX2(plasmid) | Addgene | RRID:Addgene_12260 | |
| Recombinant DNA reagent | pMD2G(plasmid) | Addgene | RRID:Addgene_12259 | |
| Recombinant DNA reagent | pAAV-MCS (plasmid) | Ruipute | Cat#: 2212E4 | |
| Recombinant DNA reagent | pAAV-RC9 (plasmid) | Ruipute | Cat#: 23011 | |
| Recombinant DNA reagent | pHelper (plasmid) | Ruipute | Cat#: 230112 | |
| Recombinant DNA reagent | pAAV-MIR4521 sponge-zsGREEn1-shRNA(plasmid) | Ruipute | Cat#: 230113 | |
| Recombinant DNA reagent | pAAV-Arg1-zsGREEn1-shRNA(plasmid) | Ruipute | Cat#: 2212E1 | |
| Recombinant DNA reagent | pAAV-ARG1-zsGREEn1-shRNA(plasmid) | Ruipute | Cat#: 2212E2 | |
| Recombinant DNA reagent | pAAV-ZsGreen1 (plasmid) | Takara | Cat#: 6231 | |
| Recombinant DNA reagent | pAAV-ZsGreen1-shRNA (plasmid) | youbio | Cat#: VT8093 | |
| Commercial assay or kit | Fluorescent In Situ Hybridization Kit | RIBOBIO | Cat#: C10910 | |
| Commercial assay or kit | RNA pull down kit | GENESEED | Cat#: P0202 | |
| Commercial assay or kit | RNA Immunoprecipitation Kit | GENESEED | Cat#: P0102 | |
| Software, algorithm | ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ | |
| Software, algorithm | Prism | GraphPad v8.0 | RRID:SCR_002798 |