Metastasis of colon cancer requires Dickkopf-2 to generate cancer cells with Paneth cell properties
Figures
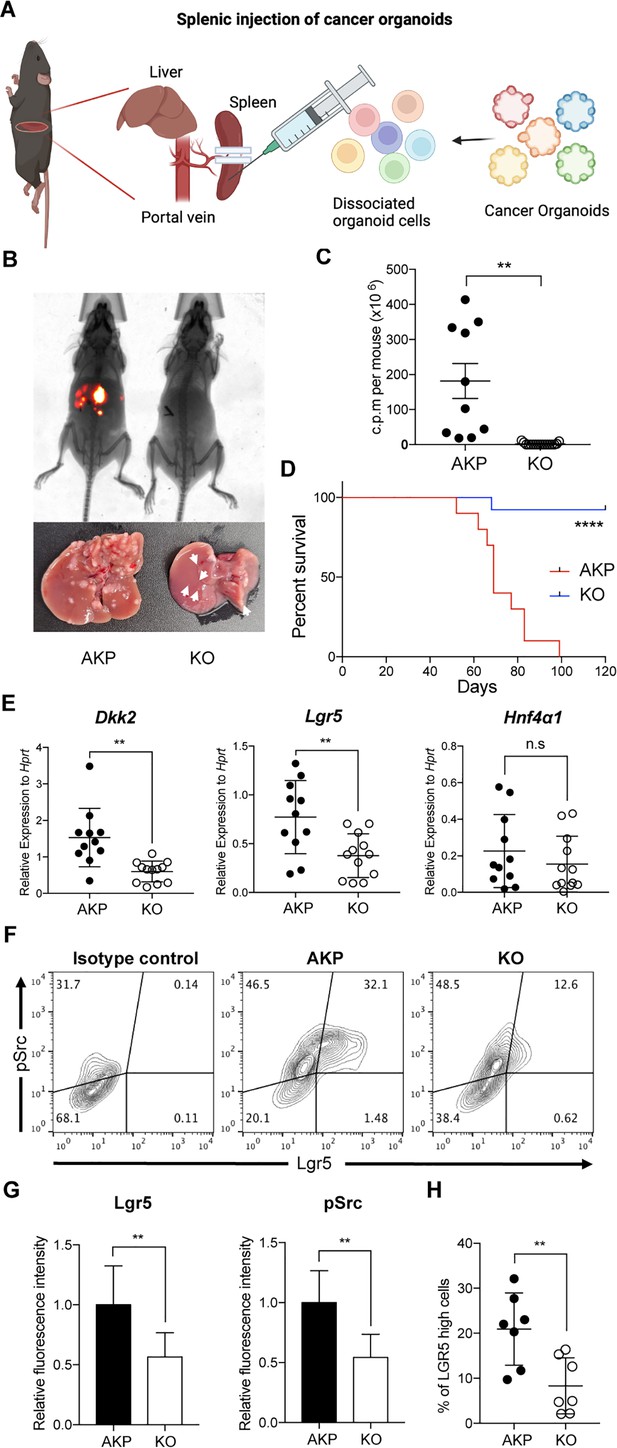
DKK2 is required for Lgr5 positive stem cells-driven liver metastasis of colorectal cancer.
(A) Isolated cells from AKP tumor organoids expressing the tdTomato reporter gene were dissociated and injected into the spleen of wild-type C57BL/6 mice. 8 weeks after the injection, metastatic tumor growth was measured by in vivo imaging analysis. (B) Representative pictures of in vivo imaging analysis. Ctrl: AKP control organoids transduced with scrambled guide RNA, KO: Dkk2 knockout AKP organoids. (C–D) Statistic analysis of liver metastasis (C) and survival (D). Ctrl (n=10), KO (n=15), n represents the number of mice. c.p.m. in (C): count per minute. (E) Quantitative gene expression analysis of Dkk2, Lgr5, and Hnf4α1 in liver metastasized colon cancer cells. Ctrl (n=11), KO (n=12), n represents the number of isolated cancer nodules with five mice per group. (F) Representative flow cytometry analysis data of c-Src phosphorylation (pSrc) and Lgr5 expression in metastasized colon cancer cells. (G) Statistic analysis of Lgr5 expression and pSrc in (E). The average of mean fluorescence intensity in control samples was set as 1 and relative fluorescence intensity was calculated. (H) Statistic analysis of the percentile of Lgr5 high (Lgr5 and pSrc double positive) cells in metastasized colon cancer in (F). Each symbol represents an individually isolated cancer nodule. n.s.=not significant, **p<0.01, ****p<0.0001; two-tailed Welch’s t-test (C, E, G, H). Error bars indicate mean ± s.d. Log-rank test (D). Results are representative of three independent experiments.
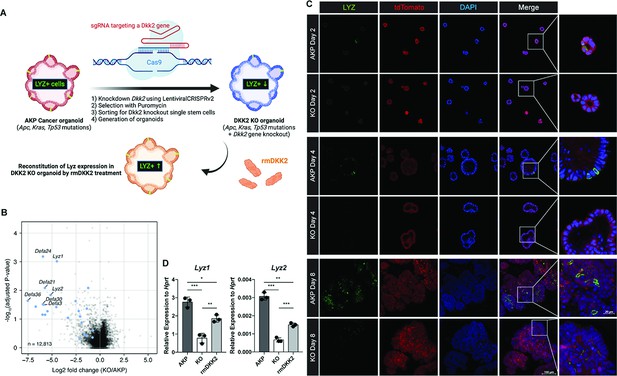
DKK2 is indispensable for the generation of cancer cells with Paneth cell properties in colon cancer organoids.
(A) A schematic diagram of the generation of KO organoids using CRISPR technique and the reconstitution of DKK2 in organoids by recombinant mouse DKK2 protein (rmDKK2) treatment. Lysozyme (LYZ) expression is highlighted for the following. (B) A volcano plot of RNA sequencing (RNA-seq) analysis comparing KO versus AKP organoids. Paneth cell marker genes are highlighted as blue circles (AKP = 3 and KO = 5 biological replicates were analyzed). (C) Confocal microscopy analysis of LYZ positive cells in AKP or KO organoids in a time-dependent manner using anti-LYZ antibody. (D) Quantitative real-time PCR analyses of Lyz1 and Lyz2 in 8 days cultured colon cancer organoids. KO organoids were cultured in the presence of 1 μg/ml of recombinant mouse DKK2 protein. *p<0.05, **p<0.01, ***p<0.001; two-tailed Welch’s t-test. Error bars indicate mean ± s.d. Results are representative of three biological replicates.
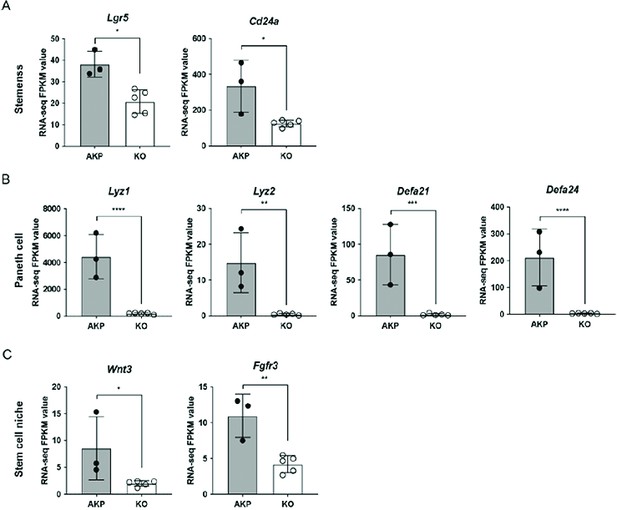
Bulk RNA sequencing (RNA-seq) analysis of Dkk2 knockout cancer organoids.
(A–C) Bulk RNA-seq analysis was performed using 8 days cultured colon cancer organoids carrying mutations in Apc, Kras, and Trp53 genes. Expression of the marker genes of stemness (A) and Paneth cells (B) are shown. Cd24a is a marker of both stemness and Paneth cells. Expression of genes related to the stem cell niche is shown in (C). *p<0.01, **p<0.05, ***p<0.01, ****p<0.001; adjusted p-values (false discovery rate) by sleuth (see Supplementary Methods). Three and five biological replicates of control (AKP) and Dkk2 KO cancer organoids were tested, respectively.
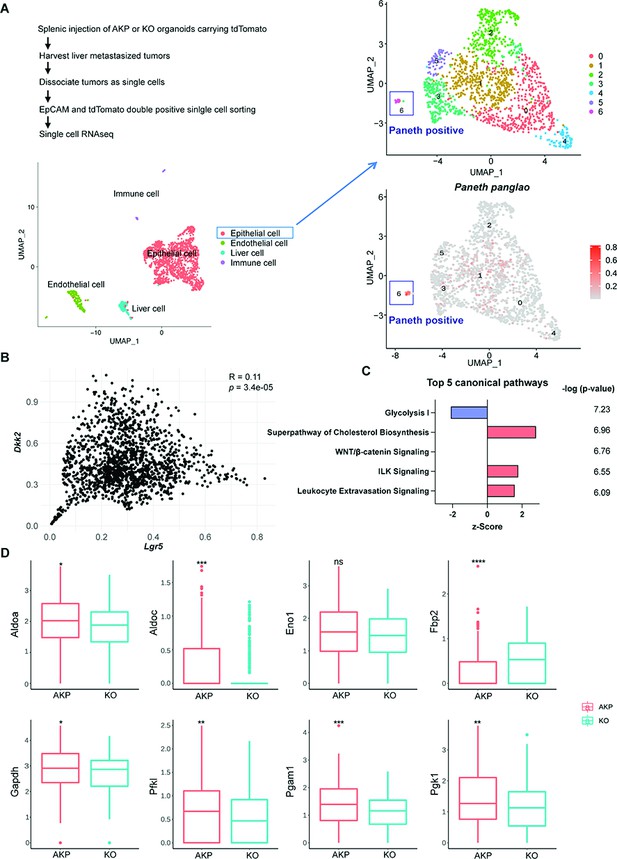
Cancer cells harboring Paneth cell properties were reduced in Dkk2 knockout metastasized colon cancer tissues in mice.
Control AKP or KO colon cancer organoids were transplanted via splenic vein as described in Figure 1. 3 weeks after transplantation, mice were sacrificed to analyze metastatic tumor growth in liver. (A) Single-cell RNA sequencing analysis (scRNA-seq) of liver metastasized colon cancer tissues. The uniform manifold approximation and projection (UMAP) plot clustered epithelial, endothelial, liver, and immune cells in metastasized cancers based on transcriptome analysis. The cancer epithelial cell cluster was sub-clustered to identify cells with Paneth cell properties (cluster 6, Paneth positive). (B) The correlation between Dkk2 and Lgr5 expression in the cluster of epithelial cells by Pearson r test. (C) Ingenuity pathway analysis (IPA)-suggested top 5 canonical pathways of the scRNA-seq data of Lgr5 positive epithelial cells in KO compared to AKP. z-Scores indicate activation or inhibition of the suggested pathways. The significance values for the pathways are calculated by the right-tailed Fisher’s exact test. (D) Box plots show expressions of the genes involved in the glycolysis I pathway in (C). ns = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; Wilcoxon signed-rank test.
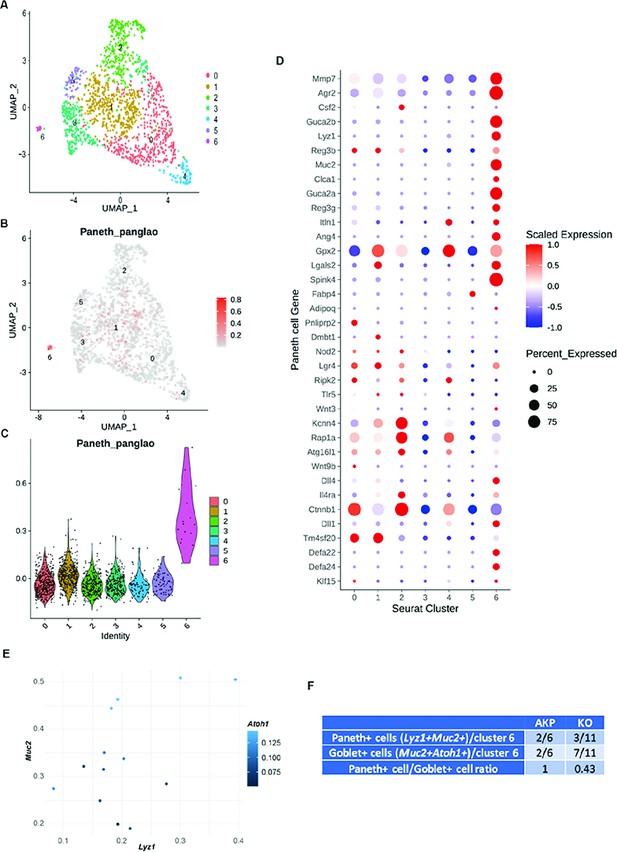
Identification of the cluster of Paneth-like cells in the single-cell RNA sequencing (scRNA-seq) data of liver metastasized murine colon cancer cells.
(A) The uniform manifold approximation and projection (UMAP) plot of tumor epithelial cells revealed seven clusters. (B) Paneth cell module score is displayed in the UMAP plot. (C) The violin plot displays the Paneth cell module score. It is the highest in cluster 6. (D) A gene expression dot plot with Paneth cell marker genes in epithelial cell clusters. (E) A scatter plot of Lyz1, Muc2, and Atoh1 expression in cluster 6 cells. (F) The ratio between cancer cells with Paneth cell properties (Paneth+) and cancer cells with goblet cell properties (Goblet+) in AKP and KO scRNA-seq data is shown.
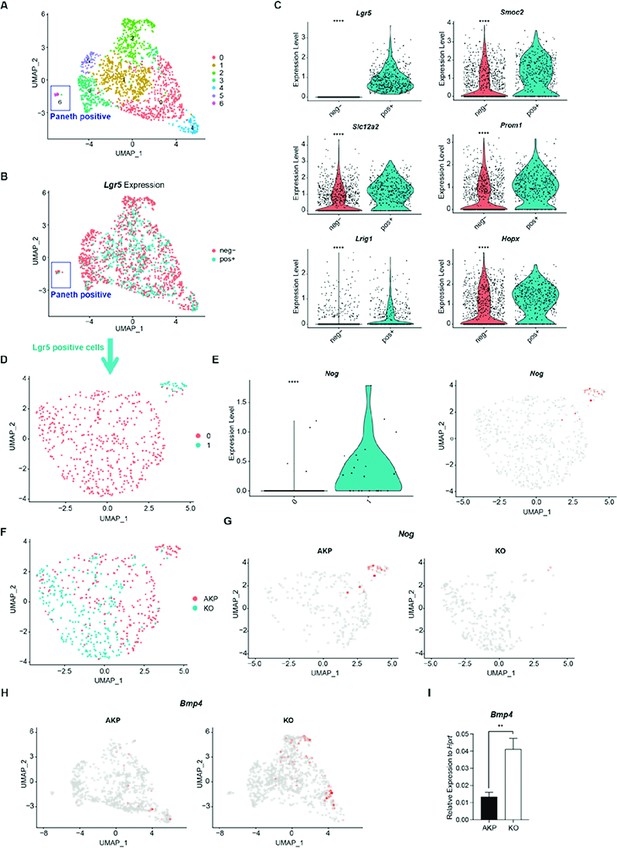
Reduced expression of Noggin (Nog) and reversed expression of Bmp4 in the single-cell RNA sequencing (scRNA-seq) of Dkk2 knockout metastasized cancer cells.
(A) The uniform manifold approximation and projection (UMAP) plot of tumor epithelial cells as described in Figure 4C. (B) Tumor epithelial cells are separated by Lgr5 expression (positive: pos+, negative: neg-). (C) Differentially expressed genes between Lgr5 positive and negative cells are presented by violin plots. ****p<0.0001; Wilcoxon signed-rank test. (D) The UMAP plot of Lgr5 positive cells revealed two clusters. (E) A violin plot displays Noggin (Nog) expression mostly detected in the cluster 1 of Lgr5 positive cells in the UMAP plot showed in (D). ****p<0.0001; Wilcoxon signed-rank test. (F) UMAP plot of Lgr5 positive cells in AKP and KO colored by red and cyan, respectively. (G) Expression of Nog in the UMAP plots of AKP and KO in (F). (H) UMAP plots of Bmp4 expression in total tumor epithelial cells of AKP and KO in the scRNA-seq data. (I) Quantitative PCR analysis of Bmp4 expression in 8 days cultured colon cancer organoids. **p<0.01; two-tailed Welch’s t-test. Error bars indicate mean ± s.d. Three biological replicates per group were tested.
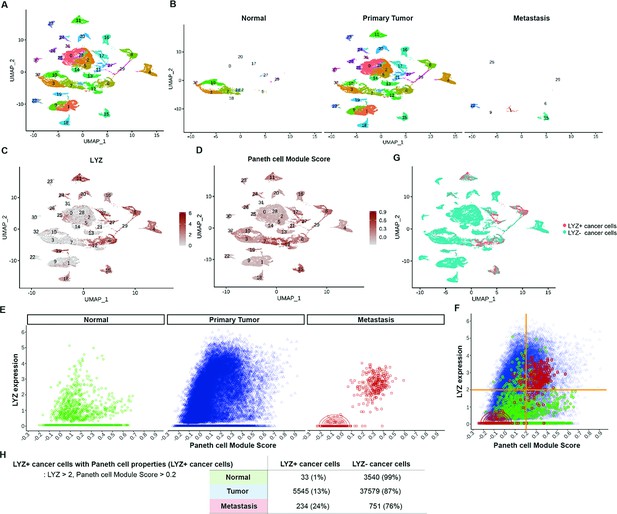
Identification of colon cancer cells harboring Paneth cell properties in humans.
Published colorectal cancer patients’ single-cell RNA sequencing (scRNA-seq) data was analyzed to identify the presence of cancer cells with Paneth cell properties (Joanito et al., 2022). (A) The uniform manifold approximation and projection (UMAP) plot of total 31 clusters. (B) Normal cells, primary tumor cells, and liver metastasized cells (metastasis) are shown in the UMAP plot clusters. (C–D) Expression levels of lysozyme (LYZ) and Paneth cell module scores are displayed in the UMPA plot. (G) Based on the analysis in (C) and (D), cancer cells harboring Paneth cell properties are indicated as red dots (LYZ+ cancer cells). (E) LYZ expression and Paneth cell module scores of single cells in normal, primary tumor, and metastasis samples are presented by dot plots. (F) Dot plots in (E) are overlayed. (H) The percentiles of cancer cells with Paneth cell properties (LYZ+ cancer cells) are shown.
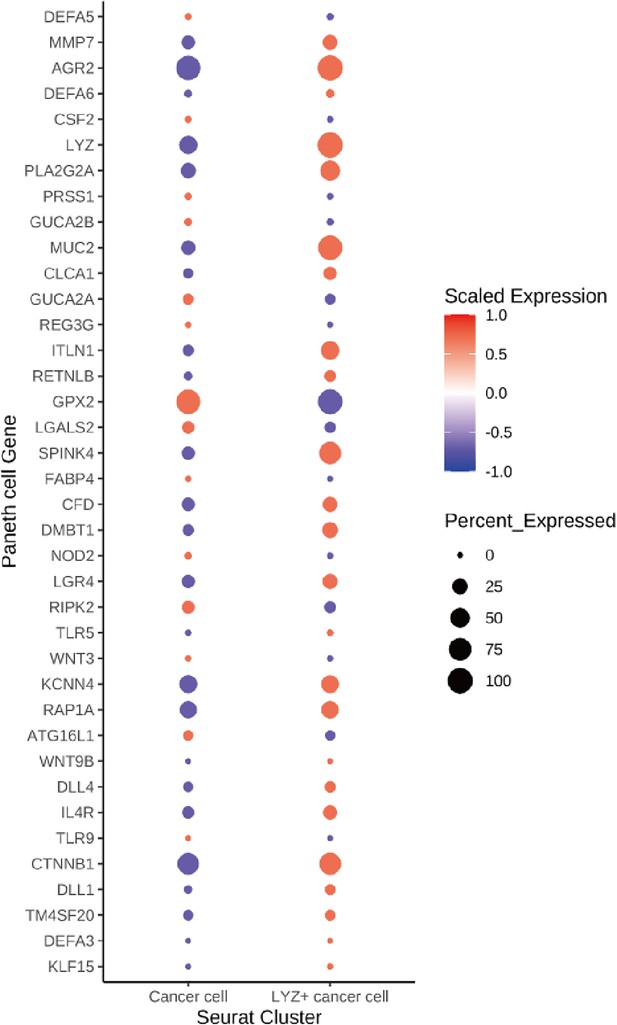
Paneth cell markers expression in colorectal cancer patients’ single-cell RNA sequencing (scRNA-seq) data.
A dot plot of Paneth cell marker genes in scRNA-seq data obtained from colorectal cancer patients. Paneth cell marker genes were retrieved from the Panglao database (PangaloDB).
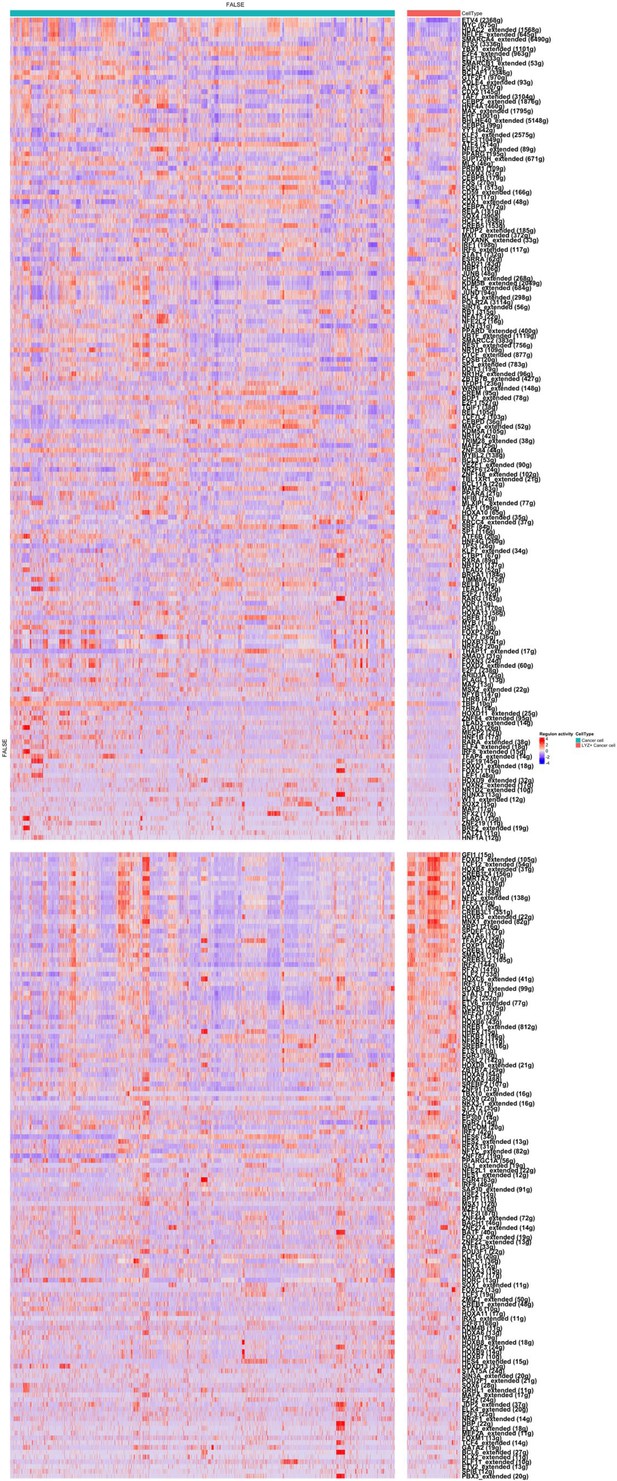
Analyses of the regulon activity of various transcription factors in lysozyme positive (LYZ+) colon cancer cells in human colon cancer single-cell RNA sequencing (scRNA-seq) data.
The z-scaled regulon activities of various transcription factors in LYZ+ cancer cells compared to LYZ- cancer cells are displayed by heatmap. The regulon activity is calculated based on the expression of transcription factors and their downstream target molecules.
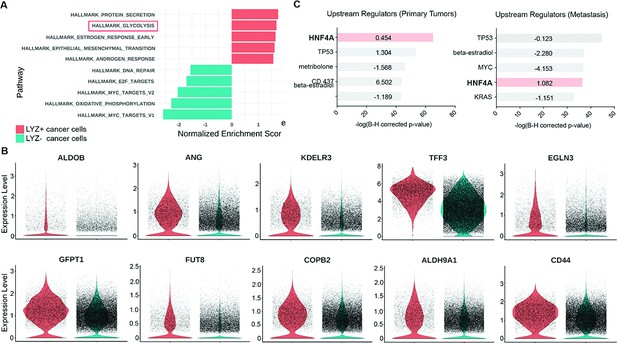
Colon cancer cells with Paneth cell properties contribute to glycolysis.
Colorectal cancer patients’ single-cell RNA sequencing (scRNA-seq) data were further analyzed by gene set enrichment analysis (GSEA) and ingenuity pathway analysis (IPA). (A) GSEA of cancer cells with Paneth cell properties (lysozyme [LYZ]+ cancer cells) compared to all other cancer cells (LYZ- cancer cells), shown in Figure 4 (G). (B) Representative gene expressions in the hallmark of glycolysis pathway are shown. (C) Upstream regulators suggested by IPA in primary tumor and metastasis are presented. Activation z-scores are indicated in the bar. IPA predicted activation or inhibition of the upstream regulators are colored as red and cyan, respectively.
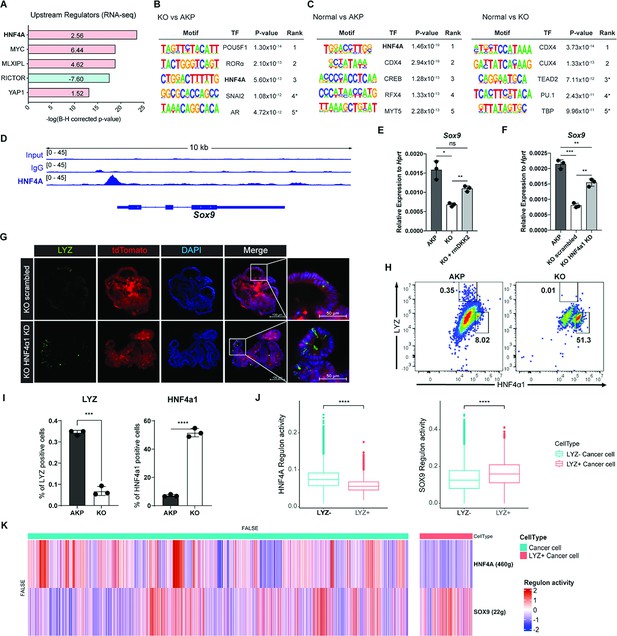
DKK2-driven reduction of HNF4α1 protein in murine colon cancer organoids promotes cancer cells with Paneth cell properties.
(A) Ingenuity pathway analysis (IPA)-suggested upstream regulators of the RNA sequencing (RNA-seq) data shown in Figure 2A. z-Scores are presented in the bar. IPA predicted activation or inhibition of the upstream regulators are colored as red and cyan, respectively. (B–C) List of top 5 transcriptional factors (TF) in the motif enrichment analysis of assay for transposase-associated chromatin using sequencing (ATAC-seq) data comparing normal colonic organoids, AKP, and KO cancer organoids. * indicates possible false positive. (D) Chromatin immunoprecipitation sequencing (ChIP-seq) analysis of knockout (KO) organoids using an anti-HNF4A antibody. (E) Quantitative expression of Sox9 in colon cancer organoids in the presence or absence of DKK2 (rmDKK2: recombinant mouse DKK2 protein added in organoid culture). (F) Analysis of Sox9 expression after knockdown HNF4α1 in KO colon cancer organoids (KO HNF4α1 KD). ns = not significant, *p<0.05, **p<0.01, ***p<0.001; two-tailed Welch’s t-test. Error bars indicate mean ± s.d. (G) Representative images of confocal microscopy analysis of Lyz-stained cancer cells with Paneth cell properties in DKK2 KO HNF4α1 KD organoids. (H–I) Control AKP or KO colon cancer organoids were transplanted via splenic vein as described in Figure 1. 3 weeks after transplantation, mice were sacrificed to analyze metastatic tumor growth in liver. Quantification of cancer cells with Paneth cell properties in metastasized tumor nodules by flow cytometry for lysozyme (LYZ) and HNF4α1. Tumor cells were initially gated by the tdTomato reporter expression. Representative images of flow cytometry are shown (H). Statistic analyses of the percentiles of LYZ positive cells (% of upper left) and HNF4α1 positive cells (% of lower right) in tumor nodules (I). **p<0.01, ****p<0.0001; two-tailed Welch’s t-test. Error bars indicate mean ± s.d. Three mice were tested per group. Data are representative of two independent experiments. (J– K) Reduced HNF4A regulation activity with enhanced SOX9 regulation activity in LYZ+ cancer cells in human colorectal cancer scRNA-seq data. Box plots represent the regulon activity of HNF4A and SOX9 in LYZ+ cancer cells. ****p<0.0001; Wilcoxon signed-rank test (J). z-Scaled regulon activities of HNF4A and SOX9 in human colon cancer cells are displayed by heatmap (K).
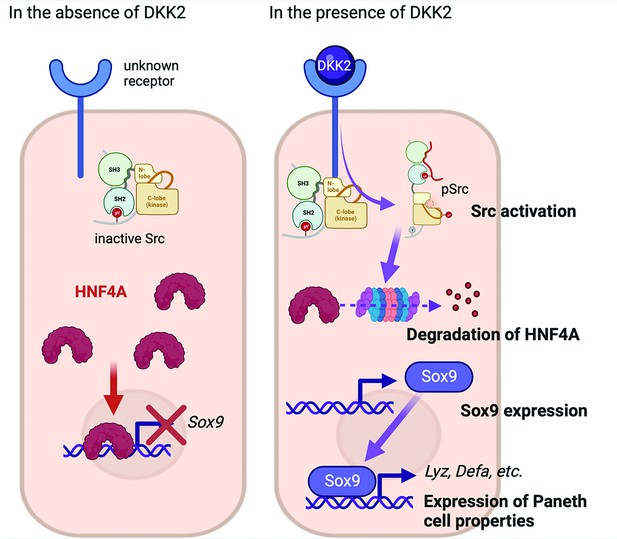
The suggested mechanism of DKK2 in the formation of colon cancer cells with Paneth cell properties.
In the absence of DKK2, Sox9 expression is inhibited by HNF4A. Our previous report has shown that DKK2 activates Src followed by degradation of HNF4A protein (Shin et al., 2021a). HNF4A deficiency leads to Sox9 expression in colon cancer cells that induces Paneth cell properties including the expression of lysozymes and defensins. Formation of cancer cells with Paneth cell properties by DKK2 contributes to the outgrowth of metastasized colon cancer cells in the liver.
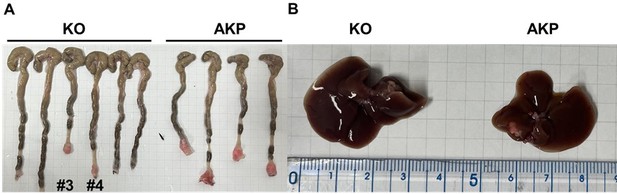
Primary tumor formation and liver metastasis by orthotopic transplantation of AKP or KO colon cancer organoids.
6-8 week-old male C57BL/6J mice were treated with 2.5% DSS dissolved in drinking water for 5 days, followed by regular water for 2 days to remove gut epithelium. After recovery with the regular water, the colon was flushed with 1000 μl of 0.1% BSA in PBS. Then, 200,000 dissociated organoid cells in 200 μl of 5% Matrigel and 0.1% BSA in PBS were instilled into the colonic luminal space. After infusion, the anal verge was sealed with Vaseline. 8 weeks after transplantation, the mice were sacrificed to measure primary tumor formation and liver metastasis.
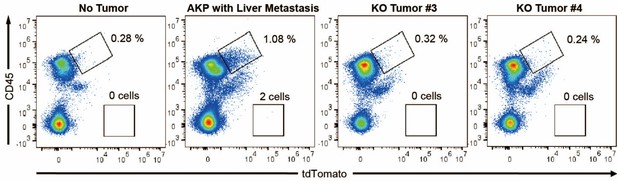
Flow cytometry analysis of tdTomato+ circulating colon tumor cells in PBMCs.
PBMCs were harvested via the portal vein after euthanasia. CD45 and tdTomato were analyzed by flow cytometry.

Pseudobulk DEG analysis confirmed the differential expression genes of interest.
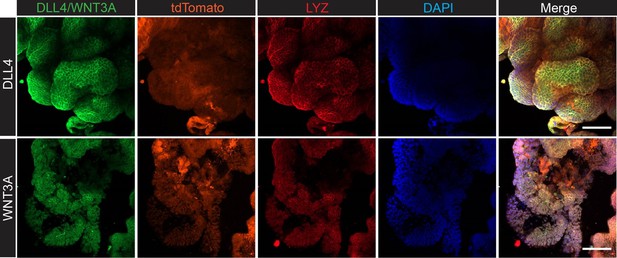
Confocal microscopy analysis for lysozyme (LYZ) and Paneth cell-derived stem cell niche factors, WNT3A and DLL4 in AKP colon cancer organoids.
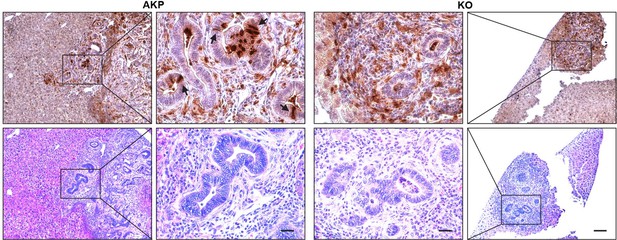
Histology of Lysozyme positive cells in metastasized tumor nodules in liver of colon cancer organoid transplanted mice.
Immunohistochemistry of Lysozyme positive Paneth-like cells cells in liver metastasized colon cancer (Upper panels, DAB staining). Identification of tumor nodules by H&E staining (lower panels, Scale bar = 100 μm). Magnified tumor nodules are shown in the 2nd and 3rd columns (Scale bar = 25 μm). Arrows indicate Lysozyme positive Paneth like cells in tumor epithelial cells. Infiltration of Lysozyme positive myeloid cells is detected in both AKP and KO tumor nodules. AKP: Control colon cancer organoids carrying mutations in Apc, Kras and Tp53 genes. KO: Dkk2 knockout colon cancer organoids
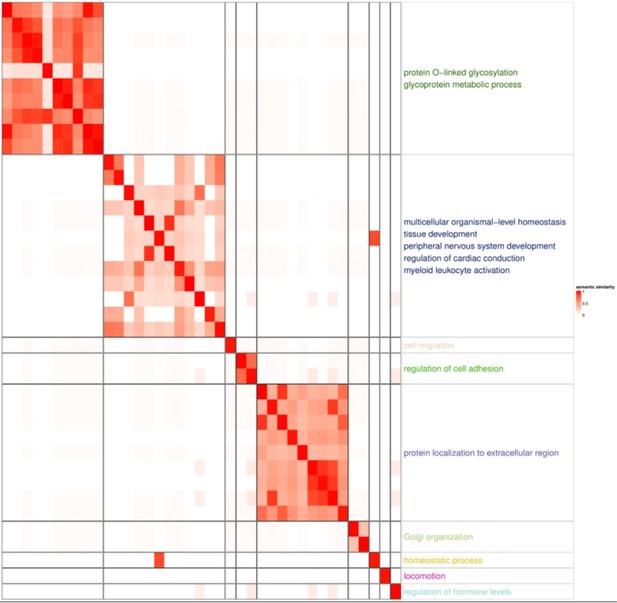
GSEA with GOBP pathway highlighted glycoprotein and protein localization to extracellular region, both of which are related Paneth cell functions.
Paneth cells secrete α-defensins, angiogenin-4, lysozyme and secretory phospholipase A2. The enriched glycoprotein process and protein localization not extracellular region reflect the characteristics of Paneth cells.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/97279/elife-97279-mdarchecklist1-v1.docx
-
Supplementary file 1
The list of primers used in quantitative real time PCR.
- https://cdn.elifesciences.org/articles/97279/elife-97279-supp1-v1.docx





