Molecular, cellular, and developmental organization of the mouse vomeronasal organ at single cell resolution
Figures

Single cell transcriptomic profile of the whole vomeronasal organ.
UMAP visualization of integrated cell-type clusters for whole-VNO single-cell RNA-seq. (B) Cell-type marker-gene normalized expression across the cell clusters. C. UMAP of cell-type clusters split by age. (D) UMAP of cell-type clusters split by sex. (E) A representative image of transcript distribution for 9 genes in a VNO slice using the Molecular Cartography platform Resolve Biosciences. Insets (a and b) show the magnified image of areas identified in the main panel. Individual cell shapes can be determined from the transcript clouds. (F) Spatial location of individual VNO cells color-coded according to cell type prediction based on the spatial transcriptomic analysis. (G) Location of cell belonging to HBC, GBC, INP, and LP cell types, respectively. Heat indicates confidence of predicted values. BL: basal lamina; MZ: marginal zone.

Age differences in gene expression.
(A) Volcano plot of gene expression differences between P14 and P56 (Wilcoxon rank sum test, FDR ≤ 0.05). (B) 16 significantly differentially expressed genes with largest positive or negative log2-fold-change values (Wilcoxon rank sum test, FDR ≤ 0.05). (C) 50 significantly enriched GO terms with largest positive or negative log2-fold-change values (GSEA Permutation testing w/ FDR ≤ 0.05).

Sex differences in gene expression.
(A) Volcano plot of gene expression differences between male and female mice (Wilcoxon rank sum test, FDR ≤ 0.05). (B) 16 significantly differentially expressed chemosensory receptors with largest positive or negative log2-fold-change values (Wilcoxon rank sum test, FDR ≤ 0.05). (C) 35 significantly enriched GO terms (GSEA Permutation testing w/ FDR ≤ 0.05).

Zonal distribution of cell types in the VNO neuroepithelia.
(A) Schematic indicating quantification of cells in the marginal, intermediate, and main zones. (B) Stacked bar-plot of cell-type proportions by VNO zone (Wilcoxon rank sum test, FDR ≤ 0.05). (C) Box plots of GBC, INP, and immature VSN cell counts by zone, across 13 slides.

Novel neuronal lineage.
(A) UMAP visualization of cell-type clusters for the neuronal lineage. (B) Expression of Gnai2 and Gnao1 in the neuronal lineage. (C) Expression of Cnga2 and Gnal in the OSN lineage. (D–E) Location of mOSNs (D) and sVSNs (E) in a VNO slice. Color indicates prediction confidence. (F) Heatmap of normalized expression for a select set of mutually differentially expressed genes between sVSNs and mature V1Rs, V2Rs, and OSNs. (G) Enriched gene ontology (GO) terms in sVSNs when compared with V1R and V2R VSNs, respectively (GSEA Permutation testing w/ FDR ≤ 0.05). (H) Box plots of normalized expression for Muc2, Obp2a, Obp2b, and Lcn3 across mature sensory neurons (Wilcoxon rank sum test, FDR ≤ 0.05).

Neuronal lineage features and differentially expressed genes in sVSNs.
(A and B) Normalized Gap43 and Stmn2 expression in the neuronal lineage. (C) Normalized. Xist expression in the neuronal lineage, split by sex. (D) Violin plot of normalized Gnai2. expression in mature V2R neurons, split by sample. (E–G) Total number of genes, counts, and percent ribosomal gene expression detected in mature neurons, split by cell type (Wilcoxon rank sum test, FDR ≤0.05). (H) Heatmap of 503 significant (Wilcoxon rank sum test, FDR ≤0.001, fold-change ≥1.5) differentially expressed genes in sVSNs.

Significantly regulated genes in sVSNs.
(A-D) Feature plots showing odor binding protein gene expression (Muc2, Obp2a, Obp2b, and Lcn3). (E–L) Top upregulated genes in sVSNs. (M–T) Top downregulated genes in sVSNs.

sVSN Pseudotime, OSN markers and GO Terms.
(A) VNO neuronal lineage cell-types in UMAP space. (B) V1R, V2R, and sVSN lineage. pseudotimes in UMAP space. (C) Volcano plot of mOSN gene expression versus all other cells. from the neuronal lineage (Wilcoxon rank sum test, FDR ≤0.05). (D) 45 significant GO terms enriched in OSNs versus all other cells (GSEA Permutation testing w/ FDR ≤0.05).

Molecular cascades separating the neuronal lineages.
(A) PCA plot of V1R and V2R pseudotime principal curves across cell types. (B) Cell density plots across pseudotime for P14 and P56 mice (Two-sided Kolmogorov-Smirnov test). (C) Heatmap showing expression in pseudotime for genes that differentially expressed between the V1R and V2R lineages. Heat indicates Z-score values. (D) Zoomed in UMAP of cell types early in the neuronal lineage. (E–I) Feature plots for select genes expressed early in the neuronal lineage. (J) UMAP of INP, iVSN, iOSN, and mOSN cell types. (K–U) Normalized expression of candidate genes for V1R/V2R/OSN lineage determination. (V) A simplified model for lineage determination by transcription factors in VNO sensory neurons.

Genes expressed during INP to immature neuron transition.
(A and B) Violin plot of normalized expression for Neurog1 (A) and Neurod1 (B) split by cell type. (C–R) Feature plots of normalized gene expression for early differentially expressed genes between V1R, V2R, and OSN lineages. (S and T) Feature plots of normalized expression for Notch1 (S) and Dll4 (T).

Receptor expression in the VNO.
(A-D) Expression level of individual receptor (total raw counts) plot against the number of cells expressing the receptor for V1R (A), V2R (B), VNO-Olfr (C), and MOE-Olfr (D). (E-H) Ranked distribution of average expression per cell for the four receptor classes. Inset pie charts show the number of cells expressing a receptor at the specified range. (I and J) Heatmaps showing the Pearson correlation coefficient of transcription factor expression among the V1Rs (I) or V2Rs (J). (K) A simplified model of transcription factor selection in mVSNs.

Receptor distribution in the VNO.
(A) Log scale rank-frequency distribution plots for V1R, V2R, VSN-OR, and OSN-OR. (B–D) Number of cells expressing (nCells) vs. total raw counts for V1R, V2R, and VSN-OR. (E–F). Rank by average count distributions for V1R, V2R, and VSN-OR.

Co-expression between vomeronasal receptors and transcription factors.
(A) Distribution density plot showing relationship between similarity of transcription factor (TF) gene expression profiles and receptor sequence similarity. (B–C) Heatmaps showing the. V1R-TF (B) and V2R-TF (C) associations. Heat shows average expression level for TF genes for a given receptor type.

Co-expression among VNO receptor classes.
(A) Shannon Indices showing the specificity of receptor expression. High values indicate more co-expressions. (B–C) Prevalence and level of receptor co-expression by age and cell- type for the V1R (B) and V2R (C) lineages, respectively. (D–F) Circos plot of genomic loci for significantly co-expressed receptor pairs in the V1R (D) V2R, (E) and across-type populations (F). (G–K) Detection of receptor gene co-expression using Molecular Cartography. Individual dots represent single molecules. Colors represent different receptor genes. DAPI stain is shown as gray. Scale bar: 10 µm.

Receptor expression statistics.
(A-C) Percent of all read counts from a receptor gene (A), number of receptor species present. per cell (B), and total counts from vomeronasal receptors (C), separated by cell type (Wilcoxon rank sum test, FDR ≤0.05). (D–G) Proportions of first, second, and third most expressed receptor gene as percent of total receptor counts, separated by cell type.
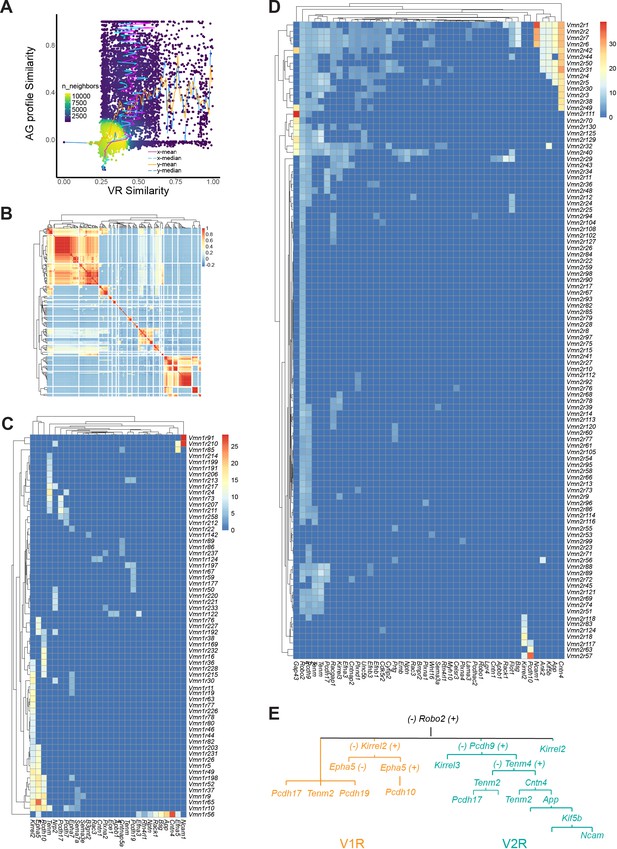
Axon guidance molecules associated with receptor genes.
(A) Distribution density plot showing relationship between similarity of axon guidance (AG) gene expression profiles and receptor sequence similarity. The distribution of receptor similarity (x-mean and x-median) remains constant over the range of AG similarity. The AG similarity (y-mean and y-median) as a function of receptor similarity shows a strong correlation at the dense part of the curve. (B) Heatmap showing the Pearson correlation coefficient among VRs in their AG expression. (C and D) Heatmaps showing the V1R-AG (C) and V2R-AG (D) associations. Heat shows average expression level for AG genes for a given receptor type. (E) A simplified model of hierarchical distribution of AG in the mVSNs.

Transcriptional determinants of axon guidance molecules for individual receptor types.
(A) Correlation heatmap between receptor types calculated from co-expressed AG and TF genes. Receptor types are color-coded. (B) 3-D heatmap showing the Jaccard Indices between AG and TF genes for each of the VRs in the dataset. (C–G) Heatmaps showing Jaccard Indices of TF-AG associations for V1R (C and D) and V2R types (E–G). The lists of TF and AG genes here are abridged from the full list to enhance visualization. (C) These receptors share Meis2 expression but different AG genes. Note that Vmn1r185 and vmn1r69 both recognize female identify pheromones. (D) Similarity and distinction of AG/TF expression for three V1R types that are located in the same genomic location and with high sequence homology. (E and F) Shared TFs and AG genes for broadly (E) and uniquely (F) expressed V2R types. (G), distinct TFs and AGGs for uniquely expressed V2R types.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus; 2 females, 2 males) | C57BL/6 | In-house breeding | ||
| Strain, strain background (Mus musculus; 4 males, 4 females) | CD-1 | In-house breeding | ||
| Commercial assay or kit | Chromium Next GEM Single Cell 3’ GEM, Library and Gel Bead Kit v3.1 | 10 X Genomics | 1000120 | |
| Commercial assay or kit | ChromiumSingle Cell 3’ GEM, Library & Gel Bead Kit v3.0 | 10 X Genomics | 1000075 | |
| Commercial assay or kit | DNase I (RNase free) | NEB | M0303 | |
| Commercial assay or kit | Papain | Calbiochem | 5125 | |
| Commercial assay or kit | DAPI | Thermo Fisher Scientific | 62247 | |
| Commercial assay or kit | L-cysteine | Calbiochem | 243005 | |
| Commercial assay or kit | BSA | Sigma-Aldrich | A8806 | |
| Commercial assay or kit | Frozen section media | Leica | 3801481 | |
| Commercial assay or kit | DRAQ5 | Invitrogen | 65-0880-96 | |
| Commercial assay or kit | HBSS | VWR | VWRL0121-0500 | |
| Commercial assay or kit | PBS | Gibco | 10010023 | |
| Commercial assay or kit | Urethane | Sigma-Aldrich | U2500 | |
| Commercial assay or kit | NovaSeq S1 | Illumina | 20012865 | |
| software, algorithm | Fiji ImageJ | Goldstein et al., 2018 | https://imagej.net/software/fiji/ | |
| Software, algorithm | QuPath v0.4.3 | Bankhead et al., 2017 | https://qupath.github.io | |
| Software, algorithm | Seurat | Hao et al., 2021 | https://satijalab.org/seurat/ | |
| Software, algorithm | kallisto | bustools | Melsted et al., 2019 | https://www.kallistobus.tools | |
| Software, algorithm | DropletUtils | Lun et al., 2019 | https://bioconductor.org/packages/release/bioc/html/DropletUtils.html | |
| Software, algorithm | Illustrator | Adobe | https://www.adobe.com/illustrator | |
| Software, algorithm | SoupX | Young and Behjati, 2020 | https://cran.r-project.org/web/packages/SoupX/index.html | |
| Software, algorithm | clustree | Zappia and Oshlack, 2018 | https://cran.r-project.org/web/packages/clustree/index.html | |
| Software, algorithm | ggplot2 | Wickham et al., 2016 | https://cran.r-project.org/web/packages/ggplot2/index.html | |
| Software, algorithm | glmGamPoi | Ahlmann-Eltze and Huber, 2021 | https://bioconductor.org/packages/release/bioc/html/glmGamPoi.html | |
| Software, algorithm | vegan | Oksanen et al., 2019 | https://cran.r-project.org/web/packages/vegan/index.html | |
| Software, algorithm | Scrublet v0.2.3 | Wolock et al., 2019 | https://github.com/swolock/scrublet | |
| Software, algorithm | reticulate | Ushey et al., 2017 | https://cran.r-project.org/web/packages/reticulate/index.html | |
| Software, algorithm | GeneOverlap | Shen, 2019 | https://bioconductor.org/packages/release/bioc/html/GeneOverlap.html | |
| Software, algorithm | circlize | Gu et al., 2014 | https://cran.r-project.org/web/packages/circlize/index.html | |
| Software, algorithm | Slingshot | Street et al., 2018 | https://www.bioconductor.org/packages/release/bioc/html/slingshot.html | |
| Software, algorithm | tradeSeq | Van den Berge et al., 2020 | https://www.bioconductor.org/packages/release/bioc/html/tradeSeq.html | |
| Software, algorithm | msigdbr | Dolgalev, 2020 | https://cran.r-project.org/web/packages/msigdbr/vignettes/msigdbr-intro.html | |
| Software, algorithm | fgsea | Korotkevich et al., 2021 | https://bioconductor.org/packages/release/bioc/html/fgsea.html | |
| Software, algorithm | Molecular Cartography | Resolve Biosciences | https://resolvebiosciences.com/ |





