Cytoneme-mediated intercellular signaling in keratinocytes is essential for epidermal remodeling in zebrafish
Figures
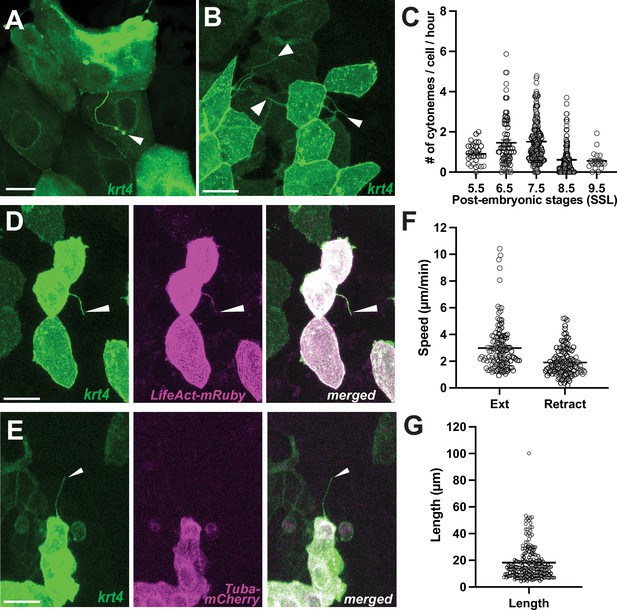
Differentiated keratinocytes extend cytoneme-like cellular protrusions.
(A) Airineme-like protrusion (arrowhead), and (B) cytoneme-like protrusions (arrowheads) labeled in krt4:palmEGFP injected keratinocytes. (C) Keratinocytes expressing krt4 extend cytoneme-like protrusions most frequently during metamorphic stages (6.5–7.5 SSL; N=457 cells, 19 larvae total). A cytoneme-like protrusion (arrowhead) labeled for (D) F-actin, revealed by LifeAct-mRuby but did not colocalize (E) with tubulin, revealed by Tuba-mCherry. (F) Keratinocyte cytonemes extend faster than they retract (N=125 cells, 4 larvae total), with an average length (G) of 18.21 µm (N=190 cells, 4 larvae total). Scale bars: 20 µm (A, B, D, E). Error bars indicate mean ± SEM.

Effect of actin inhibitor treatment on cytoneme extension.
(A) Treatment with the Cdc42 inhibitor (ML141) significantly reduced cytoneme extension frequency (p<0.0001, N=3 larvae per group). Statistical significance was assessed using the Student t test. Error bars indicate mean ± SEM.
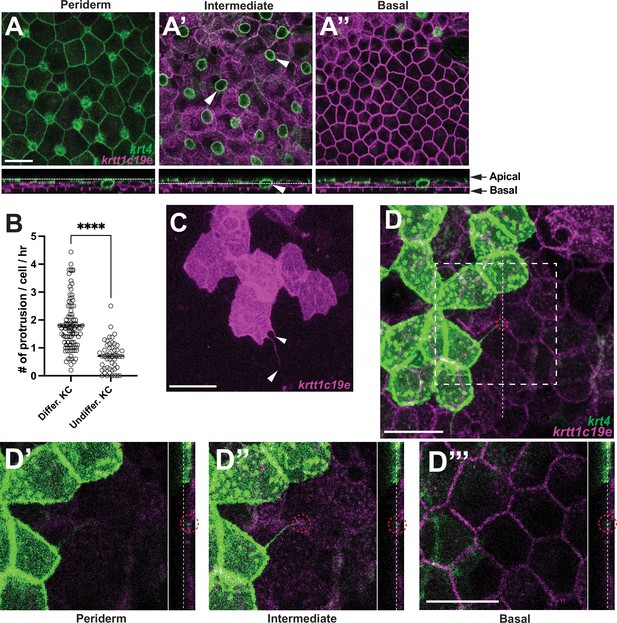
Keratinocyte cytonemes establish physical contact with underlying undifferentiated keratinocytes.
(A–A”) Postembryonic zebrafish epidermis of Tg(krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) comprises three distinct layers. (A) Expression of lyn-EGFP in krt4+ cells in the periderm layer. (A’–A”) Expression of lyn-tdTomato (pseudo-colored magenta) in krtt1c19e+ cells in (A’) the intermediate layer and (A”) the basal layer. Note that the green circular labeling in (A’, arrowheads) represents mucous-secreting goblet cells. Chang and Hwang, 2011 (B, C) Undifferentiated keratinocytes (KC) extend significantly fewer cytoneme-like protrusions (arrowheads, p<0.0001, N=49, 3 larvae; N=91 cells, 6 larvae total). (D–D”’) Cytonemes from fully differentiated keratinocytes make physical contact with undifferentiated keratinocytes in the intermediate layer (red dotted circles). Note that dotted lines indicate the layers in the cross-sectional views. Statistical significances were assessed by Student t test. Scale bars: 20 µm (A–A”, C, D–D”’). Error bars indicate mean ± SEM.
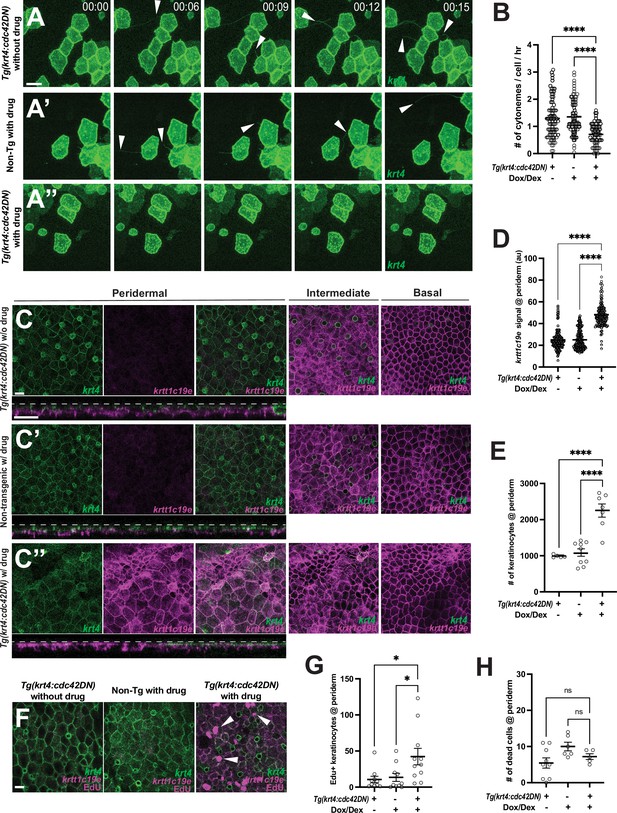
Cytoneme-mediated signaling regulates keratinocyte differentiation and proliferation.
(A–A”) Still images from time-lapse movies demonstrate that cytoneme extension (arrowheads) is inhibited in keratinocytes expressing cdc42DN. (B) Expression of cdc42DN effectively inhibits cytoneme extension (F2, 231 = 25.38, p<0.0001, N=234 cells, 10 larvae total). (C) Periderm layer of Tg(krt4:TetGBDTRE-v2a-cdc42DN;krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) without drug treatment. (C’) Periderm layer of Tg(krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) with drug treatment. (C’’) Periderm layer of Tg(krt4:TetGBDTRE-v2a-cdc42DN;krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) with drug treatment exhibits disorganization. Intermediate keratinocytes were not depleted, and basal stem cells were unaffected by the manipulation. Note that, unlike the other two controls, the epidermis was not flat, resulting in partial display of basal cells in the intermediate layer and dark areas in the basal layer. Cross-section view exhibits krtt1c19e expression at the periderm. (D) Cytoneme inhibition increases krtt1c19e expression in the periderm (F2, 387 = 226.7, p<0.0001, N=39 larvae total) and leads to an increased number of keratinocytes within the periderm (E) (F2, 18 = 27.36, p<0.0001, N=21 larvae total). (F–G) Significant increase in proliferating peridermal keratinocytes in cdc42DN expressing animals (F2, 28 = 4.888, p=0.0151, N=9 larvae). (H) The cell death rate in the periderm is not affected by cytoneme inhibition, tested with acridine orange incorporation (F2, 17 = 3.192, p=0.0666, N=20 larvae total). Statistical significances were assessed by One-way ANOVA followed by Tukey’s HSD post hoc test. Scale bars: 20 µm (A–A”, C–C”, F). Error bars indicate mean ± SEM.

Local manipulations with dominant-negative cdc42, rac1, or rhoab in peridermal keratinocytes.
(A) Mosaic expression of krt4:TetGBDTRE-cdc42DN in krt4+ keratinocytes results in a significant reduction in cytoneme extension (F2, 289=50.82, p<0.0001, N=292 cells, 9 larvae total), and (E–E’) leads to local peridermal disorganization and elevated krtt1c19e expression in krt4+ keratinocytes compared to controls shown in D and E. (B, F–F’) Similar phenotypes are observed in krt4:TetGBDTRE-rac1DN expressing peridermal keratinocytes as in krt4:TetGBDTRE-cdc42DN expressing keratinocytes (F2,239=21.43, p<0.0001, N=242 cells, 9 larvae total). (C, G–G’) However, local expression of krt4:TetGBDTRE-rhoabDN shows no effects on cytoneme extension frequency and peridermal keratinocytes (F2,237=0.08942, p=0.9145, N=240 cells, 9 larvae total). Statistical significances were assessed by One-way ANOVA followed by Tukey’s HSD post hoc test. Scale bars: 20 µm. Error bars indicate mean ± SEM.

Goblet cell counts in the zebrafish epidermis.
(A) Quantification of goblet cell numbers revealed no significant difference between two controls and experimental Tg(krt4:TetGBDTRE-v2a-cdc42DN) zebrafish (F2,20 = 2.763, p=0.0872, N=23 larvae total). (B) Quantification of goblet cell numbers showed no significant difference between two controls and experimental Tg(krtt1c19e:TetGBDTRE-v2a-SuHDN) zebrafish (F2, 30 = 3.040, p=0.0628, N=33 larvae total). Statistical significances were assessed by One-way ANOVA followed by Tukey’s HSD post hoc test.
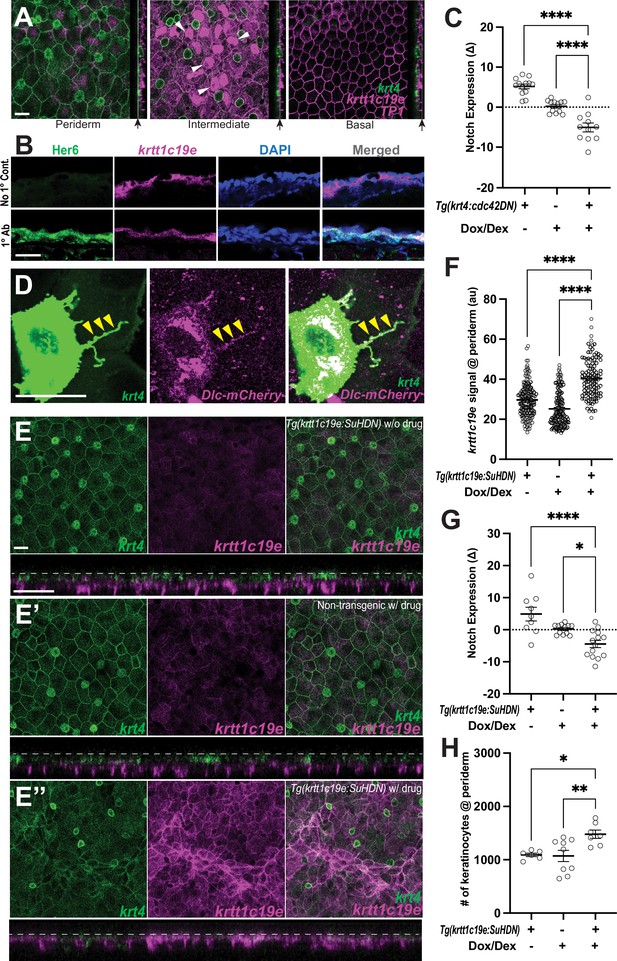
Cytonemes activate Notch in undifferentiated keratinocytes.
(A) Postembryonic zebrafish epidermis of Tg(krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato;TP1:H2BmCherry) shows Notch responsiveness in undifferentiated keratinocytes within the intermediate layer but not in the periderm or basal layer. (B) Zebrafish Her6 protein expression in krtt1c19e+intermediate keratinocytes. A cross-sectional view of Tg(krtt1c19e:tdtomato) epidermis shows that Her6 expression overlaps with the intermediate layer marker. (C) Inhibition of cytonemes reduces Notch responsiveness in undifferentiated keratinocytes (F2, 33 = 48.27, p < 0.0001, N=36 larvae total) (Figure S5C). (D) Expression of DeltaC-mCherry fusion protein along the cytoneme (yellow arrowheads). (E–E”) Notch inhibition by expressing SuHDN in undifferentiated keratinocytes results in the disorganization of periderm (E”). (E) Properly organized periderm layer of Tg(krtt1c19e:tetGBDTRE-v2a-SuHDN;krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) without drug treatment and (E’) Tg(krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) with drug treatment. (F) Notch inhibition increases krtt1c19e signal in the periderm (F2, 397 = 89.47, p < 0.0001, N=40 larvae total), (G) reduces Notch responsiveness in undifferentiated keratinocytes (F2, 32 = 13.18, p < 0.0001, N=35 larvae total), and leads to (H) an increased number of keratinocytes in the periderm (F2, 19 = 6.620, p = 0.0066, N=22 larvae total). Statistical significances were assessed by One-way ANOVA followed by Tukey’s HSD post hoc test. Scale bars, 20µm (A, C, D). Error bars indicate mean ± SEM.

Gene expressions in keratinocytes.
RT-PCR analysis revealed the endogenous expression of krt4, krtt1c19e, notch1a, notch 2, notch 3, and dlc in keratinocytes. These cells were FACS-sorted for EGFP+ cells from Tg(krt4:lyn-EGFP) and for tdTomato+ cells from Tg(krtt1c19e:tdTomato). Whole genome cDNA was used as a positive control, and reactions without a template served as negative controls.
-
Figure 4—figure supplement 1—source data 1
Original gel images for RT-PCR analysis displayed in Figure 4—figure supplement 1, with labels.
- https://cdn.elifesciences.org/articles/97400/elife-97400-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original files for RT-PCR analysis displayed in Figure 4—figure supplement 1, without labels.
- https://cdn.elifesciences.org/articles/97400/elife-97400-fig4-figsupp1-data2-v1.zip

Notch reporter (Tp1) expression.
(A) Images before and after treatment with DMSO and LY411575 (Notch inhibitor) in the Tg(Tp1:EGFP). (B) Treatment with LY411575 resulted in a significant overall reduction in Notch expression levels in the TP1 Notch reporter line compared to the DMSO control group (p<0.001, N=5 larvae per group). (C) Images before and after cytoneme inhibition through drug induction of cdc42DN-expression in krt4+ keratinocytes and direct inhibition of Notch signaling via drug induction of SuHDN expression in krtt1c19e+ keratinocytes, along with DMSO treatment. (D) The number of Tp1+ keratinocytes peaked during epidermal remodeling (6.5–7.5 SSL, N=58 larvae total) in zebrafish. (E) A significantly lower number of Tp1+ keratinocytes was observed in Tg(krt4:il17rd) or Tg(krt4:il17r1a1) compared to control (Figure 5C). Statistical significance was assessed by Student t test. Scale bars: 100 µm (A, C, E). Error bars indicate mean ± SEM.

Notch signal modifiers in undifferentiated keratinocytes.
(A–A’) Images showing lunatic fringe (lfng) expression in the intermediate layer of epidermis. Scale bars: 20 µm (A–A’).
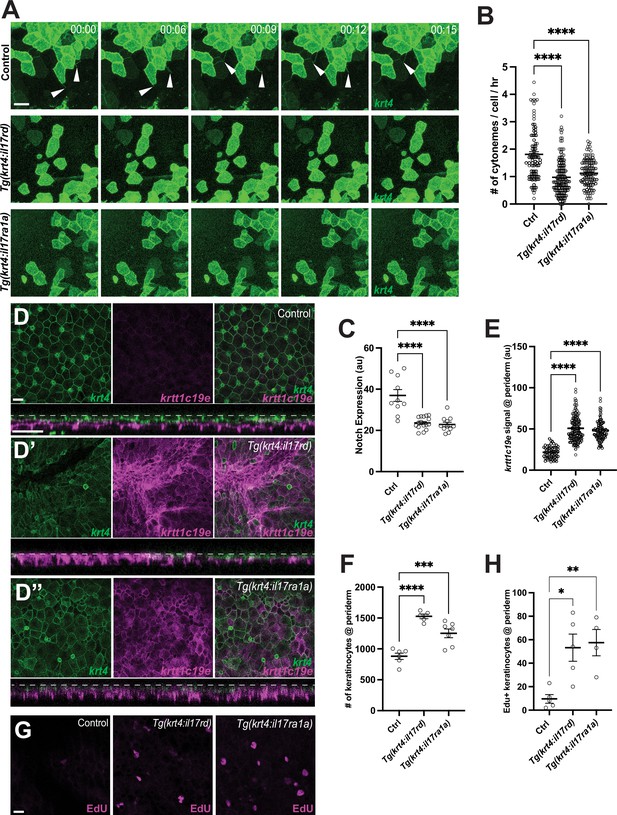
Disruption of epidermal maintenance by Interleukin-17 receptor overexpression.
Overexpression of il17rd or il17ra1a in krt4+ keratinocytes results in (A, B) a significant reduction in cytoneme extension (F2, 325 = 38.48, p < 0.0001, N=328 cells, 10 larvae total), and (C) decreased Notch responsiveness (F2, 34 = 23.16, p < 0.0001, N=37 larvae total) (Figure S5E). (D–D”) The periderm is disrupted in (D’) Tg(krt4:nVenus-v2a-il17rd;krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) and (D”) Tg(krt4:nVenus-v2a-il17ra1a;krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato) compared to the (D) properly arranged periderm of Tg(krt4:lyn-EGFP;krtt1c19e:lyn-tdTomato), resulting in (D’, cross-section) an increased krtt1c19e signal in the periderm (E) (F2, 287 = 109.7, p < 0.0001, N=29 larvae total) and (F) an increased number of keratinocytes in the periderm (F2, 15 = 27.79, p < 0.0001, N=18 larvae total). (G, H) Edu+ peridermal keratinocytes are significantly increased in il17 receptor overexpressed larvae (F2, 11=8.259, p=0.0065, N=14 larvae total). Statistical significances were assessed by One-way ANOVA followed by Tukey’s HSD post hoc test. Scale bars: 20µm (A, D, G). Error bars indicate mean ± SEM.

Overexpression of il17rd, il17ra1a, or il17a in transgenic animals.
(A) Quantitative PCR reveals that il17rd mRNA is overexpressed in Tg(krt4:il17rd) (p=0.0159, N=3 independent experiments). (B) Quantitative PCR shows il17ra1a mRNA is overexpressed in Tg(krt4:il17ra1a) (p=0.0351, N=4 independent experiments). (C) Quantitative PCR demonstrates that il17a mRNA is overexpressed in the skin of Tg(mpeg1:il17a) animals (p=0.0183, N=4 independent experiments). Statistical significances were assessed by Student t test. Error bars indicate mean ± SEM.
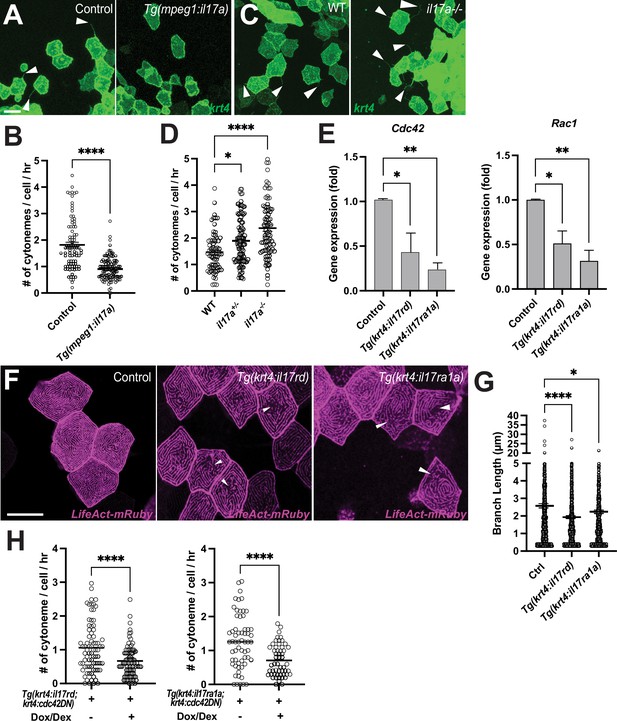
Interleukin-17 regulates keratinocyte cytonemes.
(A) Representative images of krt4:palmEGFP injected cells in control or Tg(mpeg1:il17a). (B) Significant reduction in cytoneme extension in response to overexpressed il17a ligands in the epidermis (p<0.0001, N=3 larvae per group). (C) Representative images of krt4:palmEGFP injected keratinocytes in WT or il17a-/- mutants. (D) Cytoneme extension frequency increased significantly in il17a+/- or il17a-/- mutants as compared to their wild-type siblings (F2, 247=17.85, p<0.0001, N=250 cells, 9 larvae total). (E) FACS-sorted krt4+ peridermal cells from Tg(krt4:palmEGFP; krt4:il17rd) or Tg(krt4:palmEGFP; krt4:il17ra1a) show significantly reduced Cdc42 (p=0.0341 for il17rd, p=0.01 for il17ra1a, N=3 independent experiments) and Rac1 expression (p=0.0189 for il17rd, p=0.0017 for il17ra1a, N=4 independent experiments). (F) il17rd-or il17ra1a overexpressing krt4+ keratinocytes exhibit disorganization of microridges (arrowheads), with (G) significantly reduced branch lengths of microridges in the transgenic animals (F2, 2311=11.11, p<0.0001, N=68 cells, 9 larvae total). (H) Simultaneous overexpression of cdc42DN and il17rd or il17ra1a in krt4+ keratinocytes further reduces cytoneme extension (il17rd: p<0.0001, N=164 cells, 7 larvae total, il17ra1a: p<0.0001, N=115 cells, 6 larvae total). Statistical significances were assessed by One-way ANOVA followed by Tukey’s HSD post hoc test or Student t test. Scale bars: 20 µm (A, C, F). Error bars indicate mean ± SEM.

CRISPR/Cas9 induced knockout of zebrafish il17a resulting in a premature stop codon.
(A) Comparison of Sanger sequencing chromatograms between wild-type and heterozygous il17a mutant zebrafish. The region targeted by the single guide RNA is highlighted within the black box. (B) Sanger sequencing of the il17a mutant reveals an 11 bp deletion, leading to the induction of a premature stop codon. (C) Western blotting confirmed that IL-17 protein expression was not detected in the homozygous il17a CRISPR mutants.
-
Figure 6—figure supplement 1—source data 1
Original blots for western analysis displayed in Figure 6—figure supplement 1, with labels.
- https://cdn.elifesciences.org/articles/97400/elife-97400-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Original files for western analysis displayed in Figure 6—figure supplement 1, without labels.
- https://cdn.elifesciences.org/articles/97400/elife-97400-fig6-figsupp1-data2-v1.zip

Microridge disorganization in cdc42DN or rac1DN overexpressed keratinocytes.
(A–C) Arrowheads indicate disrupted microridges in cdc42DN or rac1DN overexpressed keratinocytes. Differentiated keratinocytes (krt4+) are mosaicly labeled in the Tg(krt4:TetGBDTRE-nVenus-cdc42DN) or Tg(krt4:TetGBDTRE-nVenus-rac1DN). These transgenes were induced by administering Dox and Dex. (E) Significantly reduced branch lengths of microrides in the cdc42DN (P<0.0001, N=40 cells, 7 larvae total) or (F) rac1DN overexpressed cells (p<0.0001, N=66 cells, 6 larvae total). Statistical significances were assessed by Student t test. Scale bars: 20 µm (A, B, C). Error bars indicate mean ± SEM.

clint1 regulates epidermal maintenance via keratinocyte cytonemes.
(A–A’) clint1 mutants exhibit a disorganized periderm. (B–E) clint1 mutants show (B) a lower frequency of cytoneme extension as compared to controls (WT, cx41.8-/-) (F2, 186 = 14.55, p<0.0001, N=10 larvae total), (C) reduced Notch responsiveness (p=0.0119, N=12 mutants, N=10 controls), (D) increased krtt1c19e expression in the periderm (p<0.0001, N=13 mutants, N=6 controls), and (E) a greater number of keratinocytes within the periderm (p<0.001, N=8 mutants, N=6 controls). Statistical significances were assessed by One-way ANOVA followed by Tukey’s HSD post hoc test and Student t test. Scale bars, 20 µm (A–A’). Error bars indicate mean ± SEM.
Videos
Cytoneme extension in keratinocytes.
In cdc42DN-expressing krt4+ (green) keratinocytes (right), cytoneme extension is significantly inhibited. It is worth noting that normal filopodial and lamellipodial extension seems unaffected. In the control groups (left and middle), cytonemes actively extend. The movie was captured at 3 min intervals, and the hours and minutes elapsed are displayed in the upper right corner.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | il17rd | ZFIN | ZDB-GENE-020320–5 | Amplified from cDNA |
| Gene (Danio rerio) | il17ra1a | ZFIN | ZDB-GENE- 070705–242 | Amplified from cDNA |
| Gene (Danio rerio) | il17a/f1 | ZFIN; NCBI | GenBank accession number NM_001020787; ZDB-GENE-061031–3 | Amplified from cDNA |
| Antibody | anti-β-actin (rabbit monoclonal) | GeneTex | GTX637675; RRID:AB_3073746 | 1:5000 |
| Antibody | anti-IL-17 a/f1 (rabbit polyclonal) | KINGFISHER BIOTECH, INC | KP1239Z-100 | 1:1000 |
| Antibody | anti-Hes1 (rabbit monoclonal) | Invitrogen | MA5-32258; RRID:AB_2809544 | 1:250 |
| Antibody | Donkey anti-rabbit IgG H&L (Alexa Fluor 488) | Abcam | ab150073; RRID:AB_2636877 | 1:250 |
| Strain, strain background (Danio rerio) | WT(ABb) | PMID:26701906 | RRID:ZDB-GENO-960809-7 | Parichy Lab derivative of AB, ABwp |
| Strain, strain background (Danio rerio) | Tg(krt4:lyn-EGFP)sq18 | PMID:24400120 | RRID:ZDB-TGCONSTRCT-140415-2 | |
| Strain, strain background (Danio rerio) | Tg(krtt1c19e:lyn-tdTomato)sq16 | PMID:24400120 | RRID:ZDB-TGCONSTRCT-140424-2 | |
| Strain, strain background (Danio rerio) | Tg(krt4:TetGBDTRE-v2a-cdc42DN)ir.rt3 | This paper | N/A | Transgenic line. Maintained in Eom lab. Described in Materials and methods. |
| Strain, strain background (Danio rerio) | Tg(krtt1c19e:TetGBDTRE-v2a-SuHDN)ir.rt18 | This paper | N/A | Transgenic line. Maintained in Eom lab. Described in Materials and methods. |
| Strain, strain background (Danio rerio) | Tg(Tp1:H2BmCherry)jh11 | PMID:19595765 | RRID:ZDB-TGCONSTRCT-120419-6 | |
| Strain, strain background (Danio rerio) | Tg(Tp1:EGFP)um13 | PMID:19595765 | RRID:ZDB-TGCONSTRCT-210311-3 | |
| Strain, strain background (Danio rerio) | Tg(krt4:nVenus-v2a-il17rd)ir.rt16 | This paper | N/A | Transgenic line. Maintained in Eom lab. Described in Materials and methods. |
| Strain, strain background (Danio rerio) | Tg(krt4:il17ra1a-v2a-mCherry)ir.rt17 | This paper | N/A | Transgenic line. Maintained in Eom lab. Described in Materials and methods. |
| Strain, strain background (Danio rerio) | clint1a^hi1520Tg/+(AB) | ZIRC | RRID:ZDB-FISH-150901-2948 | |
| Strain, strain background (Danio rerio) | Tg(mpeg1:il17a-v2a-mCherry)ir.rt21 | This paper | N/A | Transgenic line. Maintained in Eom lab. Described in Materials and methods. |
| Strain, strain background (Danio rerio) | Tg(lfng:mCherry)ir.rt20 | This paper | N/A | Transgenic line. Maintained in Eom lab. Described in Materials and methods. |
| Strain, strain background (Danio rerio) | il17a-/- | This paper | N/A | CRISPR-CAS9 knock-out line. Maintained in Eom lab. Described in Materials and methods. |
| Sequence-based reagent | actb1_forward | PMID:26891128 | ENSDARG00000037746 | 5’- CATCCGTAAGGACCTGTATGCCAAC- 3’ |
| Sequence-based reagent | actb1_reverse | PMID:26891128 | ENSDARG00000037746 | 5’- AGGTTGGTCGTTCGTTTGAATCTC- 3’ |
| Sequence-based reagent | il17rd_forward | This paper | N/A | 5’- AAATGCAGCTATAAGCAGGGA –3’ |
| Sequence-based reagent | il17rd_reverse | This paper | N/A | 5’- ATGTGACTCCGAGTTTGCGA –3’ |
| Sequence-based reagent | il17ra1a_forward | This paper | N/A | 5’- TGTAAGCACTGAAGCCGATGT –3’ |
| Sequence-based reagent | il17ra1a_reverse | This paper | N/A | 5’- CACATCGAGGATGCGGAAGT –3’ |
| Sequence-based reagent | il17a/f1_forward | This paper | N/A | 5’- GATAGACGGCGTTGAGGTCC –3’ |
| Sequence-based reagent | il17a/f1_reverse | This paper | N/A | 5’- TCCACATAAGGACGAACGCA –3’ |
| Sequence-based reagent | cdc42_forward | This paper | N/A | 5’- ATACGTGGAATGCTCCGCTC –3’ |
| Sequence-based reagent | cdc42_reverse | This paper | N/A | 5’- ACGCTTCTTCTTGGGCTCTG –3’ |
| Sequence-based reagent | rac1_forward | This paper | N/A | 5’- AGGCCATAAAGTGTGTGGTCGTC –3’ |
| Sequence-based reagent | rac1_reverse | This paper | N/A | 5’- GTAGGAAAGCGGTCGAAGCCTGTC - 3’ |
| Sequence-based reagent | krt4_forward | This paper | N/A | 5’- TCAACCAGGTCTATCTCTTACTCC- 3’ |
| Sequence-based reagent | krt4_reverse | This paper | N/A | 5’- AACAGGCTCTGGTTGACAGTTAC- 3’ |
| Sequence-based reagent | krtt1c19e_forward | This paper | N/A | 5’- GCTACCACCTTCTCCAGCGGAAG- 3’ |
| Sequence-based reagent | krtt1c19e_reverse | This paper | N/A | 5’- TGCAGCACCAAATCCTCCACCAG- 3’ |
| Sequence-based reagent | dlc_forward | This paper | N/A | 5’- CGGGAATCGTCTCTTTGATAAT- 3’ |
| Sequence-based reagent | dlc_reverse | This paper | N/A | 5’- CTCACCGATAGCGAGTCTTCTT- 3’ |
| Sequence-based reagent | notch1a_forward | This paper | N/A | 5’- CGGCATCAACACCTACAACTG- 3’ |
| Sequence-based reagent | notch1a_reverse | This paper | N/A | 5’- TGGACACTCGCAGAAGAAGG- 3’ |
| Sequence-based reagent | notch2_forward | This paper | N/A | 5’- GAGTGTGTGGACCCGTTAGTATG- 3’ |
| Sequence-based reagent | notch2_reverse | This paper | N/A | 5’- GCAGGCATCATCAATGTGACAC- 3’ |
| Sequence-based reagent | notch3_forward | This paper | N/A | 5’- TCAGGATTGTTCTCTCGTTGATG- 3’ |
| Sequence-based reagent | notch3_reverse | This paper | N/A | 5’- GTGTTAAAGCATGTACCACCATTG- 3’ |
| Sequence-based reagent | il17a/f1_forward | This paper | N/A | 5’- CCTCCGCTTTCTTATGGTGAGTATAGC –3’ |
| Sequence-based reagent | il17a/f1_reverse | This paper | N/A | 5’- GGAACCACTGAATGCCAATATAGCAG –3’ |
| Recombinant DNA reagent | krt4:LifeAct-mRuby | This paper | N/A | Assembled using Gibson Assembly |
| Recombinant DNA reagent | krt4:mCherryTuba | This paper | N/A | Assembled using Gibson Assembly |
| Recombinant DNA reagent | krt4:palmEGFP | This paper | N/A | Assembled using Gibson Assembly |
| Recombinant DNA reagent | pDestTol2-CG2(Destination Vector (#395)) | Gift. PMID:17937395 | N/A | |
| Recombinant DNA reagent | pDestTol2-exorh:mCherry (Destination Vector) | Addgene | RRID:Addgene_195989 | |
| Recombinant DNA reagent | pDestTol2-exorh:EGFP (Destination Vector) | Addgene | RRID:Addgene_195983 | |
| Commercial assay or kit | MEGAshortscript T7 High Yield Transcription Kit | Invitrogen | AM1354 | |
| Commercial assay or kit | TrueCut Cas9 Protein v2 | Invitrogen | A36497 | |
| Commercial assay or kit | LR Clonase II | Invitrogen | 12538120 | |
| Commercial assay or kit | Gibson AssemblyMaster Mix | NEB | E2611L | |
| Commercial assay or kit | Gibco Stem Pro Accutase Cell Dissociation Reagent | Thermo Fisher Scientific | A1110501 | |
| Commercial assay or kit | SuperScript III CellsDirect cDNA Synthesis Kit | Fisher Scientific | 18-080-200 | |
| Commercial assay or kit | PowerUP SYBR Green Master Mix | Thermo Fisher Scientific | A25742 | |
| Commercial assay or kit | Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 594 dye | Thermo Fisher Scientific | C10339 | 500 uM |
| Chemical compound, drug | DMSO | Sigma-Aldrich | D8418 | |
| Chemical compound, drug | Doxycycline hyclate | Sigma-Aldrich | 24390-14-5 | 37.5 μM;27.5 μM |
| Chemical compound, drug | Dexamethasone | Sigma-Aldrich | 50-02-2 | 50 μM |
| Chemical compound, drug | LY411575 | Thomas Scientific | C817J63 | 3 μM |
| Chemical compound, drug | ML141 | Sigma-Aldrich | 71203-35-5 | 2 μM |
| Chemical compound, drug | Acridine Orange | Sigma-Aldrich | 65-61-2 | 2 μg/mL |
| Chemical compound, drug | 10% Formaldehyde | LabChem | LC146602 | 3.7% |
| Chemical compound, drug | TritonX-100 | CHEM-IMPEX | 01279 | 0.5%; 0.2% |
| Chemical compound, drug | 10 x PBS Buffer | Invitrogen | AM9624 | 1 x |
| Chemical compound, drug | 4% PFA | Thermo Fisher Scientific | J19943.K2 | |
| Chemical compound, drug | Tissue-Plus O.C.T. Compound | Fisher Healthcare | 23-730-571 | |
| Chemical compound, drug | Normal Goat Serum (10%) | Thermo Fisher Scientific | 50197Z | |
| Chemical compound, drug | DAPI Fluoromount-G | SouthernBiotech | 0100–20 | |
| Software, algorithm | Fiji/ImageJ | National Institutes of Health | RRID:SCR_002285 | |
| Software, algorithm | Adobe Illustrator | Adobe Inc | RRID:SCR_010279 | |
| Software, algorithm | GraphPad Prism 9 | GraphPad | RRID:SCR_002798 |





