Cell cycle and age-related modulations of mouse chromosome stiffness
Figures
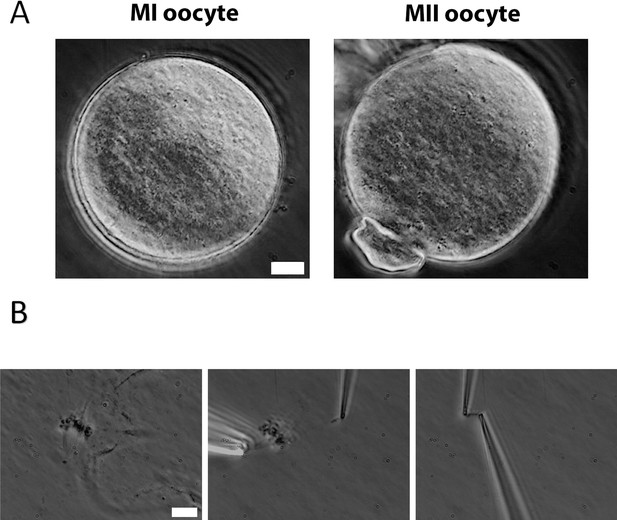
Chromosome isolation from oocytes.
(A) Oocytes after zona pellucida removal. Left panel: metaphase I (MI) oocyte. Right panel: metaphase II (MII) oocyte with visible polar body. Scale bar = 10 μm. (B) Spindle isolation process. Left panel: spindle flowing out from the oocyte after oocyte lysis. Middle panel: a chromosome being isolated from the spindle–chromosome complex. Right panel: chromosome captured between two pipettes. Scale bar = 10 μm.
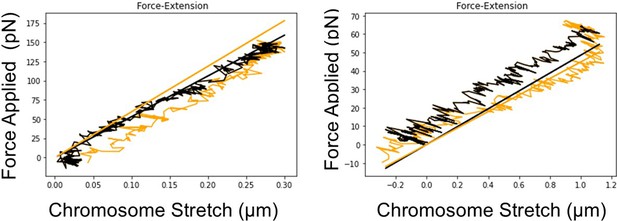
Representative images of oocyte chromosome stretching.
Left panel: metaphase I (MI) oocyte chromosome stretching. Right panel: metaphase II (MII) oocyte chromosome stretching. The black line (representing the stretching process) and the yellow line (representing the retraction process) almost overlap with each other, which indicates that oocyte chromosomes display elastic properties, and the stretching process is reversible.
-
Figure 1—figure supplement 1—source data 1
Related to Figure 1—figure supplement 1, left panel.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig1-figsupp1-data1-v1.xlsx
-
Figure 1—figure supplement 1—source data 2
Related to Figure 1—figure supplement 1, right panel.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig1-figsupp1-data2-v1.xlsx
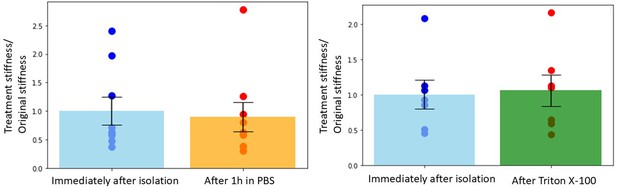
Chromosome stiffness is stable in phosphate-buffered saline (PBS) solution and resistant to Triton X-100 treatment.
Left panel: chromosome stiffness immediately after isolation (1.000 ± 0.2419, n = 9) is not significantly different from that sitting in PBS for 1 hr (0.8951 ± 0.2563, n = 9, p = 0.7697). Right panel: chromosome stiffness immediately after isolation (1.000 ± 0.2049, n = 7) shows no significant change after treatment with 0.05% Triton X-100 for 10 min (1.057 ± 0.2226, n = 7, p = 0.8527). Data are presented as mean ± SEM, with statistical analysis performed using t-test.
-
Figure 1—figure supplement 2—source data 1
Related to Figure 1—figure supplement 2, left panel.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig1-figsupp2-data1-v1.xlsx
-
Figure 1—figure supplement 2—source data 2
Related to Figure 1—figure supplement 2, right panel.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig1-figsupp2-data2-v1.xlsx
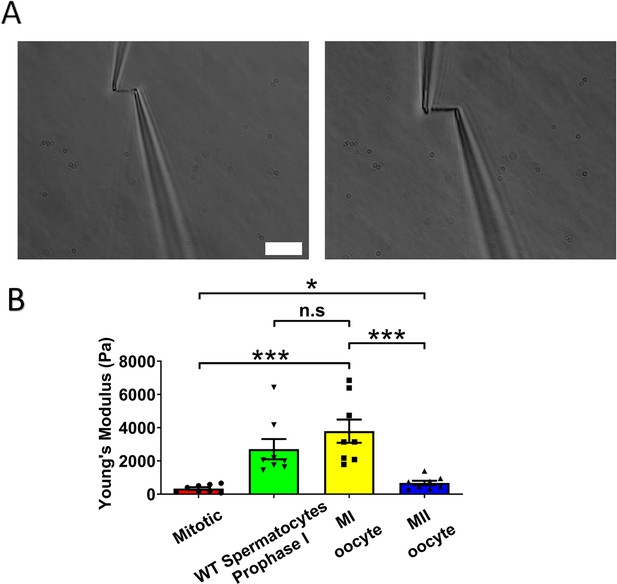
Chromosome stiffness measurement.
(A) Example images of chromosome isolation. Left: metaphase II (MII) oocyte chromosome. Right: metaphase I (MI) oocyte chromosome. Scale bar = 10 μm. (B) Chromosome stiffness comparison across different cell types: mitotic cells (n = 8), wild-type (WT) spermatocytes at prophase I (n = 8), MI oocytes (n = 8), and MII oocytes (n = 8). Young’s modulus of MI oocyte chromosomes (3790 ± 700 Pa) is much higher than that of mitotic cells (370 ± 70 Pa, p = 0.0002) and MII oocytes (670 ± 130 Pa, p = 0.0006). Data are presented as mean ± SEM. All statistical analyses were performed via t-test, n.s, non-significant, (p > 0.05), *p < 0.05, **p < 0.01 and ***p < 0.001.
-
Figure 2—source data 1
Related to Figure 2B.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig2-data1-v1.xlsx
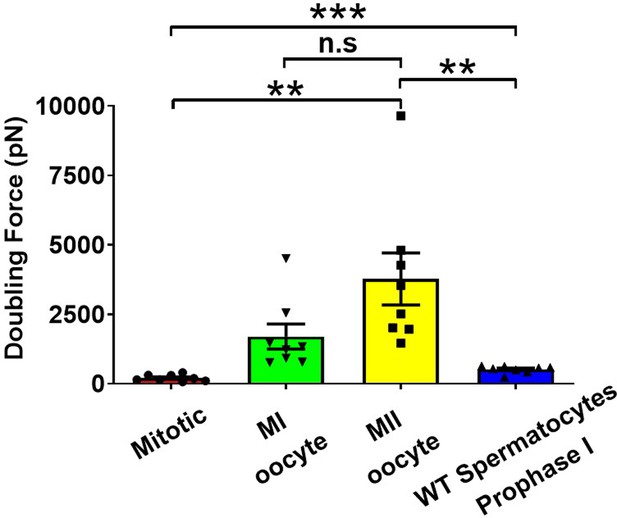
Chromosome doubling force comparison across different types of cells.
Chromosome doubling force comparison between mitotic cells (n = 8), wild-type (WT) spermatocytes at prophase I (n = 8), metaphase I (MI) oocytes (n = 8), and metaphase II (MII) oocytes (n = 8). n.s, non-significant, **p < 0.01 and ***p < 0.001.
-
Figure 2—figure supplement 1—source data 1
Related to Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig2-figsupp1-data1-v1.xlsx
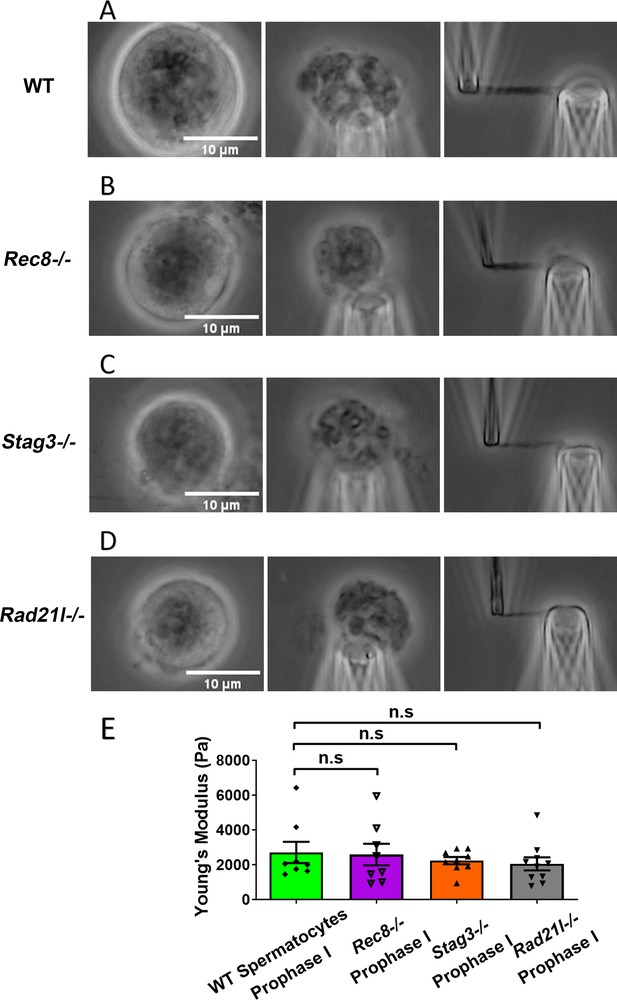
Chromosome stiffness measurement in meiosis-specific cohesin mutants.
(A) Images of chromosome isolation from wild-type (WT) spermatocytes. Scale bar = 10 μm. (B) Images of chromosome isolation from Rec8−/− spermatocytes. Scale bar = 10 μm. (C) Images of chromosome isolation from Stag3−/− spermatocytes. Scale bar = 10 μm. (D) Images of chromosome isolation from Rad21l−/− spermatocytes. Scale bar = 10 μm. (E) Chromosome stiffness comparison across various cell types: WT spermatocytes at prophase I (n = 8), Rec8−/− spermatocytes at prophase I (n = 8), Stag3−/− spermatocytes at prophase I (n = 9), and Rad21l−/− spermatocytes at prophase I (n = 10). Young’s modulus of WT spermatocyte chromosomes (2710 ± 610 Pa) is not significantly different from that of Rec8−/− spermatocyte chromosomes (2580 ± 620 Pa, p = 0.8884), Stag3−/− spermatocyte chromosomes (2240 ± 210 Pa, p = 0.4533), and Rad21l−/− spermatocyte chromosomes (2050 ± 370 Pa, p = 0.3514). Data are presented as mean ± SEM. All statistical analyses were conducted using t-test. n.s, non-significant.
-
Figure 3—source data 1
Related to Figure 3E.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig3-data1-v1.xlsx
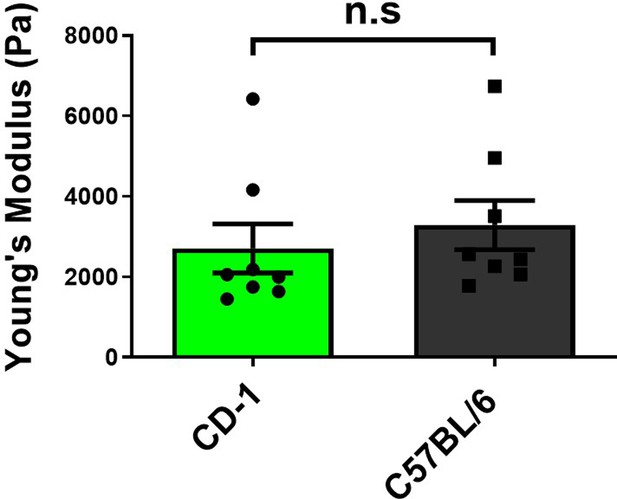
Comparison of chromosome stiffness between CD-1 and C57BL/6 mice.
Young’s modulus of chromosomes was measured and compared between CD-1 mice (n = 8) and C57BL/6 mice (n = 8). There are no significant stiffness differences between these two mouse lines (2710 ± 610 Pa in CD-1 vs. 3290 ± 610 Pa in C57BL/6J, p = 0.512). Data are presented as mean ± SEM, with statistical analysis performed using t-test. n.s, non-significant.
-
Figure 3—figure supplement 1—source data 1
Related to Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig3-figsupp1-data1-v1.xlsx
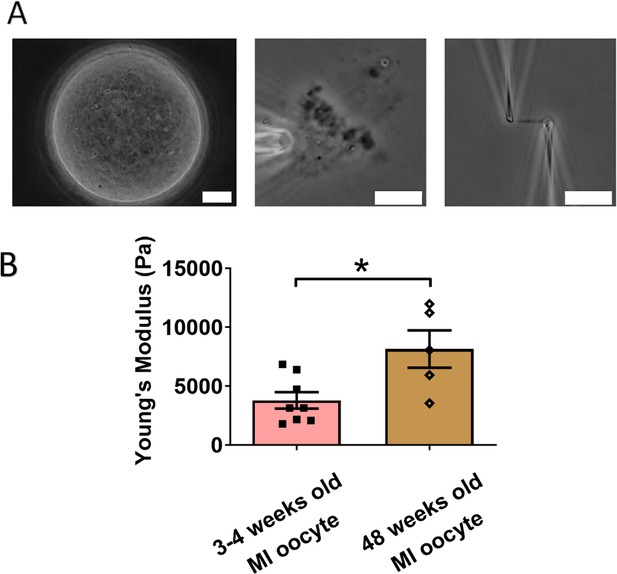
Chromosomes in aged oocytes are stiffer than those in younger oocytes.
(A) Images showing the isolation of chromosomes from an aged metaphase I (MI) oocyte. Left panel: aged MI oocyte images. Middle panel: spindle isolated from aged MI oocyte. Right panel: MI chromosome isolated from the spindle–chromosome complex. Scale bars = 10 μm. (B) Chromosome stiffness comparison between MI oocytes from 3- to 4-week-old mice (n = 8) and 48-week-old mice (n = 5). Young’s modulus of 3- to 4-week-old MI oocyte chromosomes (3790 ± 700 Pa) is significantly lower than that of 48-week-old MI oocyte chromosomes (8150 ± 1590 Pa, p = 0.0150). Data are presented as mean ± SEM and statistical analysis was performed using t-test. *p < 0.05.
-
Figure 4—source data 1
Related to Figure 4B.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig4-data1-v1.xlsx
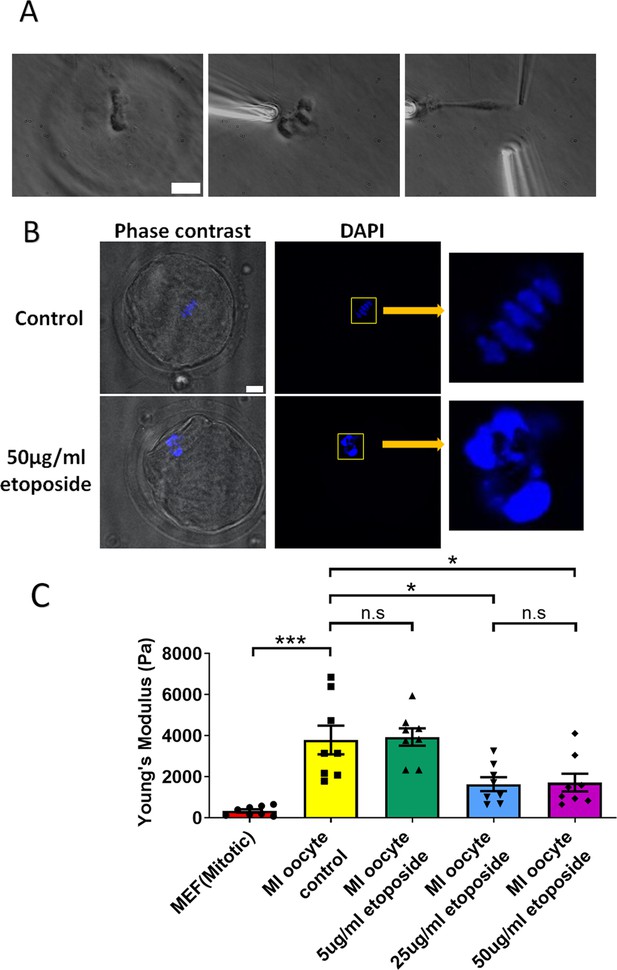
Etoposide treatment reduces chromosome stiffness.
(A) Images of chromosome isolation from metaphase I (MI) oocyte treated with 50 μg/ml etoposide. Left panel: a spindle after cell lysis. Middle panel: a spindle captured with pipettes. Right panel: chromosome isolation. Scale bar = 10 μm. (B) 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) staining of control and 50 μg/ml etoposide-treated MI oocytes. Scale bar = 10 μm. (C) Chromosome stiffness comparison between mitotic cells (n = 8), control MI oocytes (n = 8), 5 μg/ml etoposide-treated MI oocytes (n = 8), 25 μg/ml etoposide-treated MI oocytes (n = 8), and 50 μg/ml etoposide-treated MI oocyte (n = 8). Young’s modulus of control MI oocyte chromosomes (3790 ± 700 Pa) did not differ significantly from that of 5 μg/ml etoposide-treated MI oocyte chromosomes (3930 ± 400 Pa, p = 0.8624). However, it was significantly higher than that of 25 μg/ml etoposide-treated MI oocyte chromosomes (1640 ± 340 Pa, p = 0.015) and 50 μg/ml etoposide-treated MI oocyte chromosomes (1710 ± 430 Pa, p = 0.0245). Data are presented as mean ± SEM, with statistical analysis conducted using t-test. n.s, non-significant, *p < 0.05 and ***p < 0.001.
-
Figure 5—source data 1
Related to Figure 5C.
- https://cdn.elifesciences.org/articles/97403/elife-97403-fig5-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus) | CD-1 | Charles River Laboratories, Wilmington, MA | RRID:IMSR_CRL:022 | |
| Strain, strain background (M. musculus) | C57BL/6 | This paper and papers from Dr. Philip W. Jordan’s lab | RRID:IMSR_JAX:000664 | Mutant mouse lines maintained in Dr. Jordan’s lab |
| Cell line (M. musculus) | Mouse embryonic fibroblasts | Dr. John Marko’s lab | ATCC | |
| Chemical compound, drug | Etoposide | Cayman Chemical | Item No. 12092 | 50 μg/ml |
| Chemical compound, drug | M2 medium | Sigma-Aldrich | M7167-100ML | |
| Chemical compound, drug | M16 medium | Sigma-Aldrich | M7292-50ML | |
| Chemical compound, drug | EmbryoMax Acidic Tyrodes Solution | Sigma-Aldrich | MR-004-D | |
| Chemical compound, drug | IBMX | Sigma-Aldrich | I5879 | 100 μM |
| Chemical compound, drug | Triton X-100 | US Biological | 9002-93-1 | 0.05% |
| Chemical compound, drug | DAPI | Sigma-Aldrich | D9542 | 1:10,000 |
| Software, algorithm | LabVIEW | RRID:SCR_014325 | ||
| Software, algorithm | ImageJ | RRID:SCR_003070 | ||
| Software, algorithm | NIS-Elements | RRID:SCR_014329 | ||
| Software, algorithm | GraphPad Prism | RRID:SCR_002798 |





