Negative regulation of APC/C activation by MAPK-mediated attenuation of Cdc20Slp1 under stress
Figures
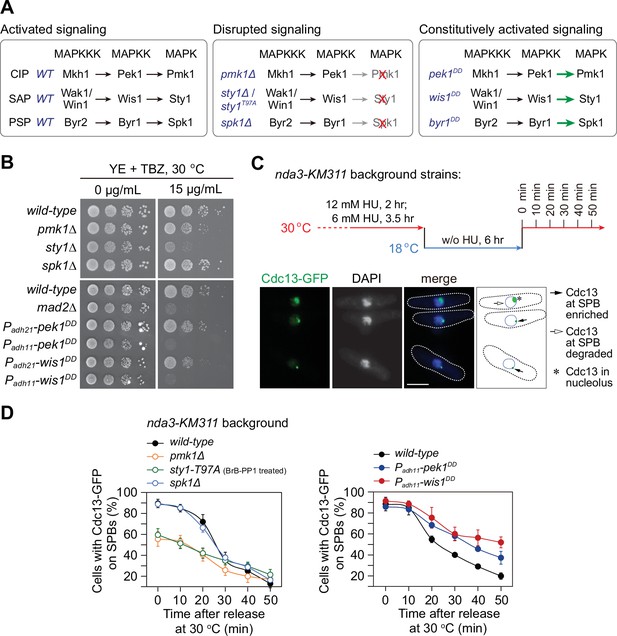
pmk1∆ and sty1-T97A mutants display spindle checkpoint activation defects, while pek1DD and wis1DD mutants are defective in checkpoint silencing.
(A) Schematic of core modules of three Schizosaccharomyces pombe mitogen-activated protein kinase (MAPK) signaling pathways. Each cascade consists of three core kinases (MAP kinase kinase kinase (MAPKKK), MAP kinase kinase (MAPKK), and MAPK). CIP, the cell integrity pathway; SAP, the stress-activated pathway; PSP, pheromone signaling pathway. These pathways can be disrupted by pmk1Δ, sty1Δ, or (adenosine triphosphate) ATP analog-sensitive mutation sty1-T97A or spk1Δ, and constitutively activated by mutations in MAPKKs Pek1 (pek1DD, pek1-S234D;T238D), Wis1 (wis1DD, wis1-S469D;T473D), or Byr1 (byr1DD, byr1-S214D;T218D), respectively. (B) Serial dilution assay on thiabendazole (TBZ) sensitivity of all MAPK deletion mutants and pek1DD- or wis1DD-overexpressing mutants. mad2∆ is a positive control. Note Padh11 is a stronger version of Padh21 promoter. (C) Schematic depiction of the experiment design for time-course analyses on spindle assembly checkpoint (SAC) or anaphase-promoting complex/cyclosome (APC/C) activation. nda3-KM311 cells carrying Cdc13-GFP were grown, synchronized with hydroxyurea (HU) and treated at 18°C for 6 hr to activate SAC, and finally shifted back to the permissive temperature 30°C. Samples were collected at 10 min intervals and subjected to microscopy analyses. Example pictures of cells with Cdc13-GFP signals enriched or disappeared at spindle pole bodies (SPBs) are shown. Scale bar, 5 μm. (D) Time-course analyses of SAC activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes. sty1-T97A was inactivated by 5 μM 3-BrB-PP1. For each time point, ≥300 cells were counted for every sample. The experiment was repeated ≥3 times and error bars indicate mean ± standard deviation.
-
Figure 1—source data 1
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 1D.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-data1-v1.xlsx
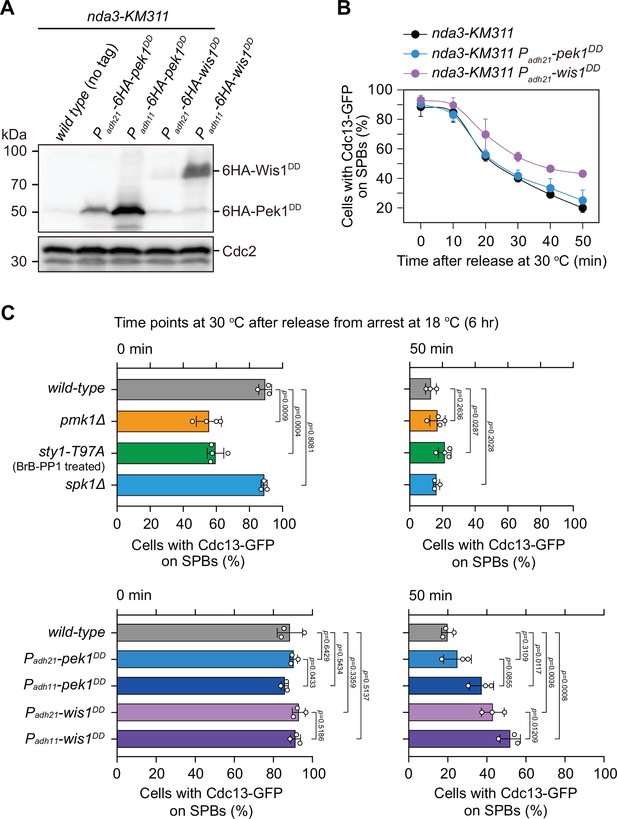
Examination of protein levels of overexpressed Pek1DD and Wis1DD and analyses of spindle checkpoint inactivation efficiency in Padh21-pek1DD and Padh21-wis1DD mutants.
(A) Immunoblot analysis of the constitutively overexpressed Pek1DD and Wis1DD. Cells with indicated genotypes were grown at the permissive temperature for nda3-KM311 (30°C) to mid-log phase and then arrested at 18°C for 6 hr. Protein samples were prepared and immunoblotting was performed with anti-HA and anti-Cdc2 antibodies to detect total 6HA-Pek1DD, 6HA-Wis1DD, and Cdc2, respectively. Note Padh11 is a stronger version of Padh21 promoter. (B) Time-course analyses of spindle assembly checkpoint (SAC) activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes. Compared with Padh11-pek1DD or Padh11-wis1DD mutants, Padh21-pek1DD or Padh21-wis1DD mutants showed weaker defect in spindle checkpoint inactivation efficiency, respectively. n ≥ 3. For each time point, ≥300 cells were counted for every sample. (C) Quantification of spindle checkpoint inactivation rate in mitogen-activated protein kinase (MAPK)-deficient or constitutively active mutants at 0 and 50 min after release at 30°C from nda3-mediated arrest. Cells of indicated strains bearing Cdc13-GFP were grown at 30°C to mid-log phase and arrested at 18°C for 6 hr, and then released at 30°C. For each time point, ≥300 cells were counted for every sample. Data from time points of 0 and 50 min after release were subjected to statistical analysis. Error bars indicate mean ± standard deviation of three independent experiments. p values were calculated against wild-type cells.
-
Figure 1—figure supplement 1—source data 1
Uncropped blots for Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp1-data1-v1.pdf
-
Figure 1—figure supplement 1—source data 2
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 1—figure supplement 1B, C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp1-data2-v1.xlsx
-
Figure 1—figure supplement 1—source data 3
Full raw unedited blot (anti-HA) for Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp1-data3-v1.zip
-
Figure 1—figure supplement 1—source data 4
Full raw unedited blot (Cdc2) for Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp1-data4-v1.zip
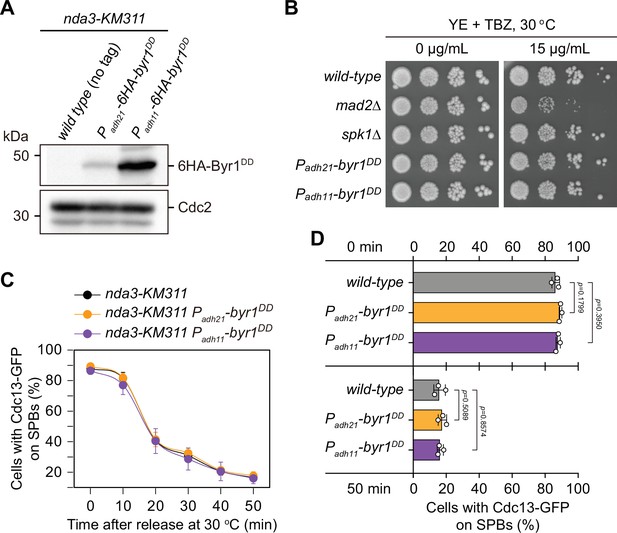
Characterization of possible effect of overexpressed Byr1DD on spindle checkpoint activation and inactivation.
(A) Immunoblot analysis of the constitutively overexpressed Byr1DD. nda3-KM311 strains with indicated genotypes were first grown at 30°C to mid-log phase and then arrested at 18°C for 6 hr. Protein samples were prepared and immunoblotting was performed with anti-HA and anti-Cdc2 antibodies to detect total 6HA-Byr1DD and Cdc2, respectively. Note Padh11 is a stronger version of Padh21 promoter. (B) Serial dilution assay on thiabendazole (TBZ) sensitivity of Byr1DD-overexpressing mutants. mad2∆ served as a positive control. Note that neither Padh21-6HA-byr1DD nor Padh11-6HA-byr1DD mutants were sensitive to TBZ. (C) Time-course analyses of spindle assembly checkpoint (SAC) activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes. Cells of indicated strains were grown at 30°C to mid-log phase and arrested at 18°C for 6 hr, and then released at 30°C. For each time point, ≥300 cells were counted for every sample. The experiment was repeated three times and error bars indicate mean ± standard deviation. (D) Quantification of spindle checkpoint inactivation rate in byr1DD mutants at 0 and 50 min after release at 30°C from nda3-mediated arrest. Data from time points of 0 and 50 min collected in (C) were subjected to statistical analysis. Error bars indicate mean ± standard deviation of three independent experiments. p values were calculated against wild-type cells.
-
Figure 1—figure supplement 2—source data 1
Uncropped blots for Figure 1—figure supplement 2A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp2-data1-v1.pdf
-
Figure 1—figure supplement 2—source data 2
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 1—figure supplement 2C, D.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp2-data2-v1.xlsx
-
Figure 1—figure supplement 2—source data 3
Full raw unedited blot (anti-HA) for Figure 1—figure supplement 2A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp2-data3-v1.zip
-
Figure 1—figure supplement 2—source data 4
Full raw unedited blot (Cdc2) for Figure 1—figure supplement 2A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp2-data4-v1.zip
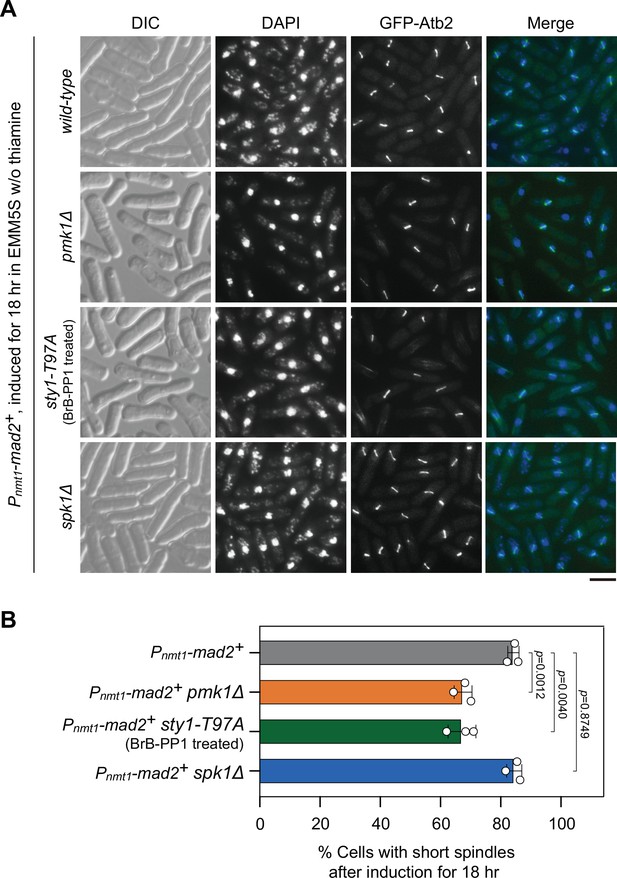
Analyses of metaphase arrest efficiency in pmk1Δ, sty1-T97A, and spk1Δ mutants upon Mad2 overexpression.
Cultures of Pnmt1-mad2+ GFP-atb2+ strains with indicated genotypes were induced in EMM5S liquid media in the absence of thiamine for 18 hr, and cells were collected, fixed, and 4',6-diamidino-2-phenylindole (DAPI) stained, and cells with short spindles were scored. For sty1-T97A strain, 5 μM 3-BrB-PP1 was added at 16 hr after induction. (A) Representative images of cells from indicated strains with GFP-Atb2-labeled spindles. DIC, differential interference contrast microscopy. Scale bar, 5 μm. (B) Quantitative analyses of cells with short spindles in response to Mad2 overexpression. The experiment was repeated three times with >280 cells counted for each strain, and the data are plotted as mean ± standard deviation. Two-tailed unpaired t-test was used to derive p values.
-
Figure 1—figure supplement 3—source data 1
Raw data of quantitative analyses of cells with short spindles in response to Mad2 overexpression for Figure 1—figure supplement 3B.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig1-figsupp3-data1-v1.xlsx
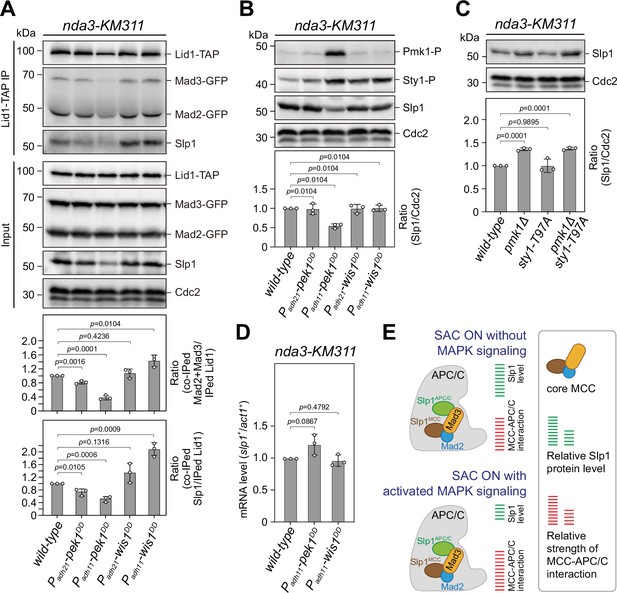
Upon spindle checkpoint activation, Slp1Cdc20 levels are reduced in pek1DD and the mitotic checkpoint complex (MCC)–anaphase-promoting complex/cyclosome (APC/C) association is enhanced in wis1DD cells compared to wild-type cells.
(A) Co-immunoprecipitation analysis on MCC–APC/C association. Cells with indicated genotypes were grown at 30°C to mid-log phase and arrested at 18°C for 6 hr. Lid1-TAP was immunoprecipitated and associated Mad2, Mad3, and Slp1Cdc20 were detected by immunoblotting. The amount of co-immunoprecipitated Mad2, Mad3, and Slp1Cdc20 was quantified by being normalized to those of total immunoprecipitated Lid1 in each sample, with the relative ratio between Mad2-GFP plus Mad3-GFP or Slp1Cdc20 and Lid1-TAP in wild-type sample set as 1.0. Blots are representative of three independent experiments. p values were calculated against wild-type cells. (B, C) Immunoblot analysis of Slp1Cdc20 abundance in nda3-KM311 cells treated at 18°C for 6 hr. Slp1Cdc20 levels were quantified with the relative ratio between Slp1Cdc20 and Cdc2 in wild-type strain set as 1.0. Phosphorylated Pmk1 (Pmk1-P) or phosphorylated Sty1 (Sty1-P) in (A) were detected using anti-phospho p42/44 and anti-phospho p38 antibodies and represents activated cell integrity pathway (CIP) or stress-activated pathway (SAP) signaling, respectively. sty1-T97A was inactivated by 5 μM 3-BrB-PP1. Blots shown are the representative of three independent experiments. p values were calculated against wild-type cells. (D) Real time quantitative PCR (RT-qPCR) analysis of mRNA levels of slp1+. Cells with indicated genotypes were grown and treated as in (A–C) before RNA extraction. The relative fold-change (slp1+/act1+) in mRNA expression was calculated with that in wild-type cells being normalized to 1.0. Note mRNA level of slp1+ in pek1DD mutant is not decreased. Error bars indicate mean ± standard deviation of three independent experiments. Two-tailed unpaired t-test was used to derive p values. (E) Schematic summary of the negative effect of activated CIP and SAP signaling on APC/C activation based on primary phenotype characterization of pmk1Δ, sty1-T97A, pek1DD, and wis1DD mutants.
-
Figure 2—source data 1
Uncropped blots for Figure 2A–C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig2-data1-v1.pdf
-
Figure 2—source data 2
Raw data of co-immunoprecipitation rate of Mad2/Mad3/Slp1, Slp1 level measurement, and RT-qPCR for Figure 2A–D.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Raw unedited blots for Figure 2.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig2-data3-v1.zip
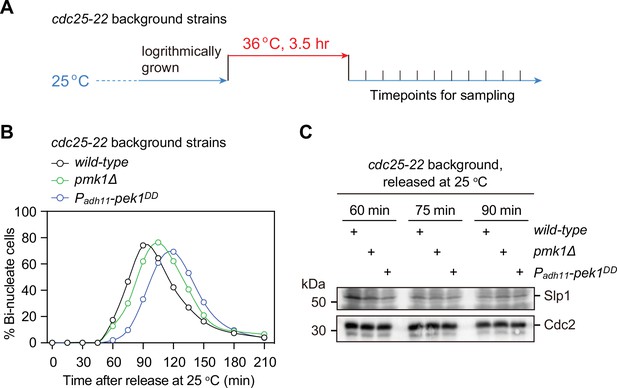
Examination of protein levels of Slp1Cdc20 in normally growing mitotic pmk1Δ orpek1DD cells.
(A) Schematic depiction of the arrest-and-release experiment design. For cdc25-22 background strains, cells were grown at 25°C and then arrested at the G2/M transition by shifting to 36°C for 3.5 hr. The cultures were released by shifting back to 25°C, and aliquots were taken at different time intervals and were fixed and stained with DAPI to monitor cell-cycle progression. (B) The percentages of binucleated cells were counted based on DAPI staining for each time point after release at 25°C. (C) Samples of indicated strains taken at 60, 75, and 90 min after being shifted back from 36 to 25°C were subjected to immunoblotting with anti-Slp1 and anti-PSTAIR antibodies to detect total Slp1 and Cdc2, respectively. Blots shown are representative of two independent biological replicates.
-
Figure 2—figure supplement 1—source data 1
Uncropped blots for Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig2-figsupp1-data1-v1.pdf
-
Figure 2—figure supplement 1—source data 2
Raw data of time-course analyses of arrest-and-release of cdc25-22 mutants for Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig2-figsupp1-data2-v1.xlsx
-
Figure 2—figure supplement 1—source data 3
Full raw unedited blot (Slp1) for Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig2-figsupp1-data3-v1.zip
-
Figure 2—figure supplement 1—source data 4
Full raw unedited blot (Cdc2) for Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig2-figsupp1-data4-v1.zip
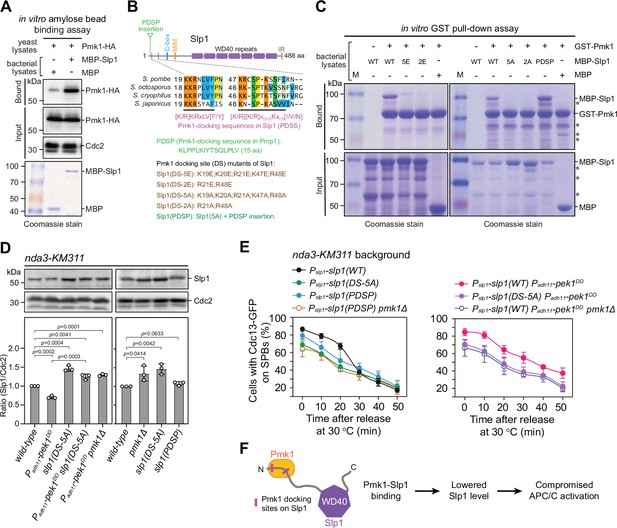
Pmk1 directly binds Slp1Cdc20 to attenuate its levels upon spindle checkpoint activation.
(A) In vitro binding assay using bacterially expressed MBP-Slp1Cdc20 and yeast lysates prepared from nda3-KM311 pmk1-HA-6His cells arrested at 18°C for 6 hr. Note that weak band detected in MBP sample was due to unspecific background binding to amylose beads. Coomassie blue staining shows inputs for MBP and MBP-Slp1Cdc20. (B) Schematic depiction of the S. pombe Slp1Cdc20 protein structure with the positions of two confirmed basic-residue patches mediating Slp1Cdc20–Pmk1 association indicated by pink bars. Alignment highlights the conservation of basic-residue patches within four Schizosaccharomyces species. The deduced Pmk1-docking motifs and different versions of motif mutations are shown. MIM, Mad2-interaction motif; IR, isoleucine–arginine tail. (C) In vitro GST pull-down assays with bacterially expressed recombinant GST-Pmk1 and MBP fusions of wild-type Slp1Cdc20 or Slp1Cdc20 mutants harboring Pmk1-docking motif mutations. An aliquot of the same amount of MBP-Slp1Cdc20 as that added in each GST pull-down reaction was immobilized by amylose resin as the input control. Asterisks indicate unspecific or degraded protein bands. (D) Immunoblot analysis of Slp1Cdc20 abundance in nda3-KM311 cells with indicated genotypes treated at 18°C for 6 hr. Slp1Cdc20 levels were quantified as in Figure 2B, C. The experiment was repeated three times. The mean value for each sample was calculated, and p values were calculated against wild-type or pek1DD cells. (E) Time-course analyses of spindle assembly checkpoint (SAC) activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes. For each time point, ≥300 cells were counted for every sample. The experiment was repeated three times and the mean value for each sample was calculated as in Figure 1D. (F) Schematic summarizing the negative effect of Pmk1–Slp1Cdc20 association on Slp1Cdc20 abundance and anaphase-promoting complex/cyclosome (APC/C) activation.
-
Figure 3—source data 1
Uncropped blots for Figure 3A, C, D.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig3-data1-v1.pdf
-
Figure 3—source data 2
Raw data of Slp1 level measurement and time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 3D, E.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig3-data2-v1.xlsx
-
Figure 3—source data 3
Raw unedited blots for Figure 3.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig3-data3-v1.zip
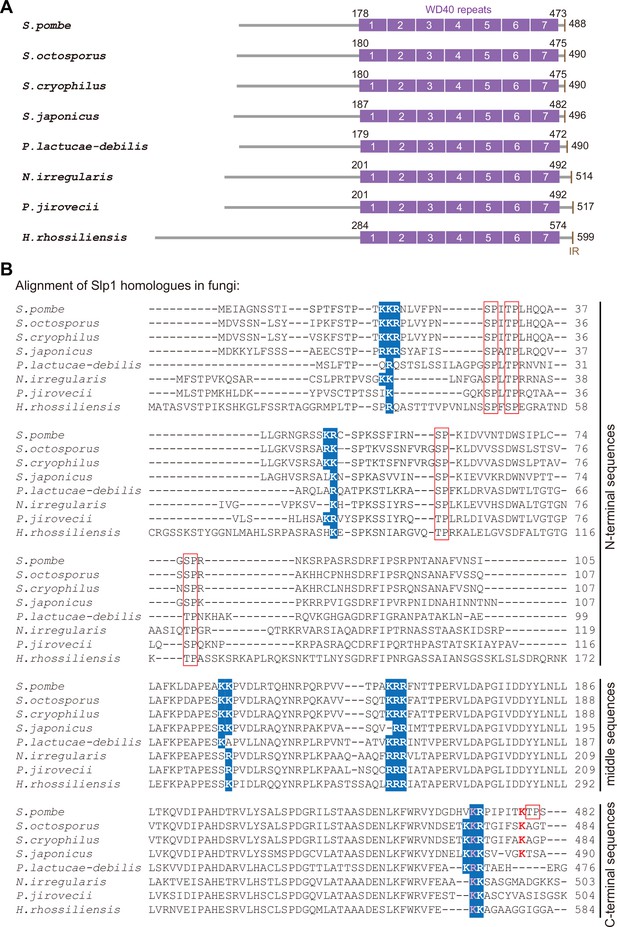
Conservation of basic-residue patches and phosphorylation or ubiquitylation sites within Slp1 homologs in fungi species.
(A) Schematic depiction of the S. pombe (Schizosaccharomyces pombe) Slp1 protein with its homologs in seven other fungi species: Schizosaccharomyces octosporus, Schizosaccharomyces cryophilus, Schizosaccharomyces japonicas, Protomyces lactucae-debilis, Neolecta irregularis, Pneumocystis jirovecii, and Hirsutella rhossiliensis. Positions of amino acids corresponding to seven WD40 repeats and IR (isoleucine–arginine) motif are indicated. (B) Local sequence alignment performed with S. pombe Slp1 and its homolog sequences from seven other fungi species. Five potential basic-residue patches within Slp1 and their conserved positions in other species are highlighted in blue. Phosphorylated or ubiquitylated residues detected by mass spectrometry in S. pombe Slp1 together with their corresponding residues in homologs from seven other fungi species are labeled with red frames or red letters, respectively. Lysine 472 (K472) in S. pombe Slp1 and its corresponding residues in homologs from seven other fungi species are also indicated in pink. Note that only aligned local sequences (N-terminal, middle and C-terminal portions) of Slp1 homologs harboring the basic-residue patches are shown.
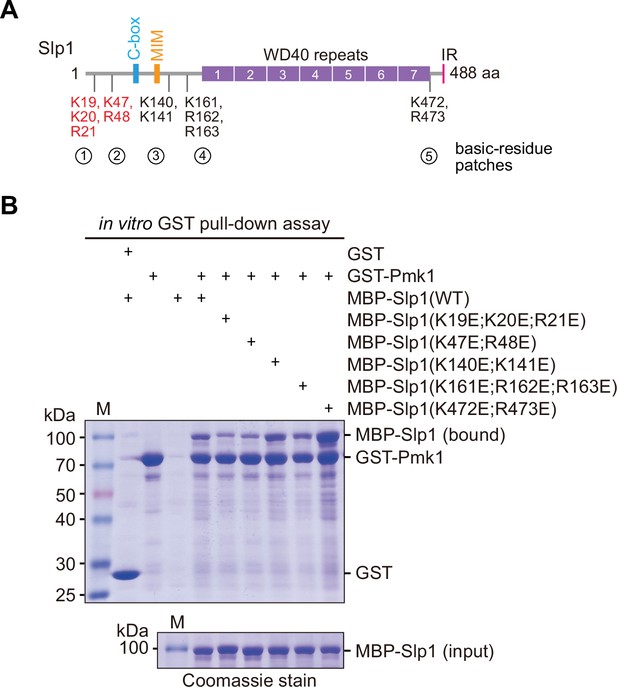
Screen for basic-residue patches in Slp1 mediating its direct interaction with Pmk1 by in vitro binding assay.
(A) Schematic depiction of the S. pombe Slp1 protein with five basic-residue patches potentially required for Slp1–Pmk1 association indicated, and two confirmed interaction-mediating patches highlighted in red. MIM, Mad2-interaction motif; IR, isoleucine–arginine tail. (B) In vitro GST pull-down assays were performed with bacterially expressed GST-Pmk1 and MBP-Slp1 with wild-type Slp1 or mutants harboring lysine/arginine (K/R) to glutamic acid (E) mutations of basic-residue patches within Slp1. Proteins bound on GST beads were separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and visualized by Coomassie blue staining. Note that only Slp1 harboring clustered mutations K19E, K20E and R21E, or K47E and R48E within two most N-terminal basic-residue patches strongly compromised interaction between Slp1 and Pmk1.
-
Figure 3—figure supplement 2—source data 1
Uncropped gels for Figure 3—figure supplement 2B.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig3-figsupp2-data1-v1.pdf
-
Figure 3—figure supplement 2—source data 2
Full raw unedited Coomassie gel (bead-bound GST-Pmk1 and MBP-Slp1) for Figure 3—figure supplement 2B.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig3-figsupp2-data2-v1.zip
-
Figure 3—figure supplement 2—source data 3
Full raw unedited Coomassie gel (MBP-Slp1 input) for Figure 3—figure supplement 2B.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig3-figsupp2-data3-v1.zip
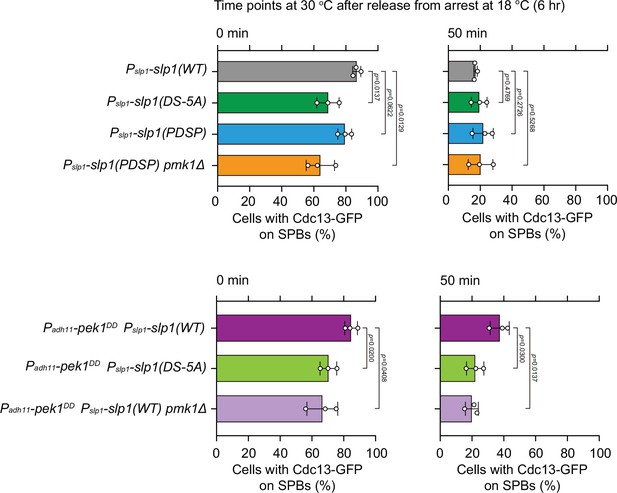
Quantification of spindle checkpoint inactivation rate in slp1 mutants with Pmk1-docking site mutations at 0 and 50 min after release at 30°C from nda3-mediated arrest.
Cells of indicated strains bearing Cdc13-GFP were grown at 30°C to mid-log phase and arrested at 18°C for 6 hr, and then released at 30°C. For each time point, ≥300 cells were counted for every sample. Data from time points of 0 and 50 min after release in Figure 3E were subjected to statistical analysis. Error bars indicate mean ± standard deviation of three independent experiments. p values were calculated against wild-type cells.
-
Figure 3—figure supplement 3—source data 1
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig3-figsupp3-data1-v1.xlsx
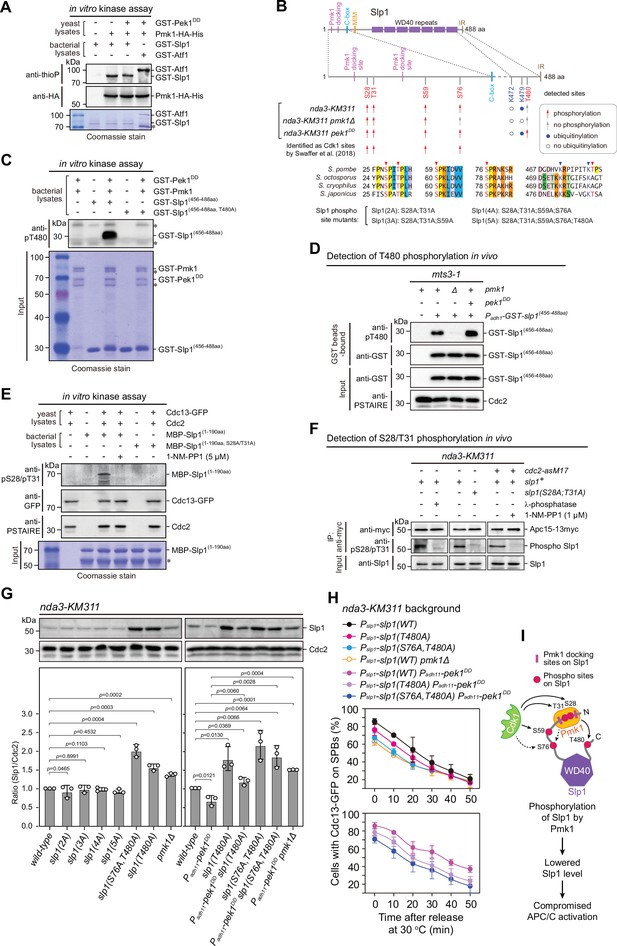
Pmk1 synergizes with Cdk1 to phosphorylate and reduce Slp1Cdc20 abundance.
(A) Non-radioactive in vitro phosphorylation assays with bacterially expressed recombinant GST-Slp1Cdc20 and Pmk1-HA-His purified from yeast cells. The incorporation of the thiophosphate group was determined using anti-thiophosphate ester antibodies (anti-thioP) as indicative of phosphorylation. Note that the presence or absence of GST-Pek1DD does not affect Slp1Cdc20 phosphorylation efficiency. A known Pmk1 substrate Atf1 was used as a positive control (Atf1-P). Asterisks indicate bands corresponding to unspecific or likely degraded proteins. (B) Summary of mass spectrometry data on Slp1Cdc20 phosphorylation and ubiquitylation in vivo in nda3-KM311-arrested cells. Red arrows and filled blue circles denote all detected phosphorylated or ubiquitylated sites, respectively, while gray arrows and unfilled circles indicate the absence of phosphorylation or ubiquitylation in some of these sites, respectively. Alignment highlights the conservation of most of the detected phosphorylation and ubiquitylation sites within four Schizosaccharomyces species. (C) In vitro phosphorylation assays with bacterially expressed recombinant GST-fusion of Slp1Cdc20 fragment (456–488aa), GST-Pmk1 and GST-Pek1DD. The reactions were blotted with pT480 antibodies. Asterisks indicate bands corresponding to unspecific or likely degraded proteins. (D) Immunoblot detection of Slp1Cdc20 phosphorylation at T480 in vivo. GST-slp1(456–488aa) was purified from mts3-1 cells with indicated genotypes arrested at 36°C for 3.5 hr, and detected with anti-GST and anti-pThr480 antibodies. Note that pThr480 is absent in pmk1∆ cells, and enhanced in pek1DD cells relative to that in wild-type cells. (E) In vitro phosphorylation assays with bacterially expressed recombinant MBP-fusion of Slp1Cdc20 fragment (1–190aa) and Cdc13 (cyclin B)-containing Cdk1 complexes purified from metaphase-arrested nda3-KM311 yeast cells. 1-NM-PP1 was added as inhibitor for analog-sensitive Cdc2-as. The reactions were blotted with pS28/pT31 antibodies. Asterisks indicate bands corresponding to unspecific or likely degraded proteins. (F) Immunoblot detection of Slp1Cdc20 phosphorylation at S28/T31 in vivo. Apc15-13myc was immunoprecipitated from nda3-KM311 cells treated at 18°C for 6 hr and the samples were blotted with pS28/pT31 antibodies. One IP sample from wild-type background was treated with λ-phosphatase. For cdc2-asM17 cells, 1-NM-PP1 was added to inactivate Cdc2 during culturing. (G) Immunoblot analysis of Slp1Cdc20 abundance in nda3-KM311 cells treated at 18°C for 6 hr. Slp1Cdc20 levels were quantified as in Figure 2B, C. The experiment was repeated three times. The mean value for each sample was calculated, and p values were calculated against wild-type or pek1DD cells. (H) Time-course analyses of spindle assembly checkpoint (SAC) activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes. For each time point, ≥300 cells were counted for every sample. The experiment was repeated three times and the mean value and p value for each sample were calculated as in Figure 1D. (I) Schematic summarizing the negative effect of Slp1Cdc20 phosphorylation by Pmk1 on its abundance and anaphase-promoting complex/cyclosome (APC/C) activation.
-
Figure 4—source data 1
Uncropped blots for Figure 4A, C–G.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-data1-v1.pdf
-
Figure 4—source data 2
Raw data of Slp1 level measurement and time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 4G, H.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Raw unedited blots for Figure 4.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-data3-v1.zip
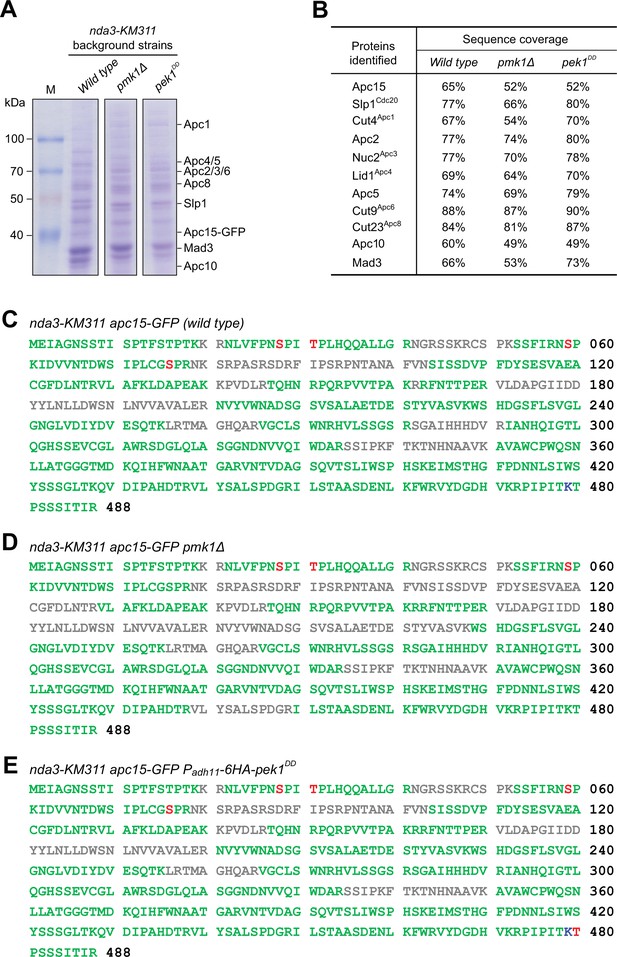
Identification of in vivo phosphorylated or ubiquitylated residues in Slp1.
(A, B) Anaphase-promoting complex/cyclosome (APC/C) subunits were isolated by immunoprecipitation of Apc15-GFP from nda3-KM311 cells arrested at metaphase by being treated at 18°C for 6 hr. Purified proteins were analyzed by SDS–PAGE and mass spectrometry. Coomassie blue-stained protein gels after SDS–PAGE are shown (A). APC/C subunits or related proteins identified by Apc15-GFP purifications followed by mass spectrometry are listed, and percentages of peptide sequence coverage for each protein are indicated (B). (C–E) Slp1 sequences retrieved from three purifications in indicated strains with peptide sequence coverage (green), phosphorylated serine or threonine (red), and ubiquitylated lysine (blue). Sequences not covered after mass spectrometry analysis are in gray.
-
Figure 4—figure supplement 1—source data 1
Uncropped gels for Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp1-data1-v1.pdf
-
Figure 4—figure supplement 1—source data 2
Full raw unedited Coomassie gel (wild-type) for Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp1-data2-v1.zip
-
Figure 4—figure supplement 1—source data 3
Full raw unedited Coomassie gel (pmk1Δ, pek1DD) for Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp1-data3-v1.zip
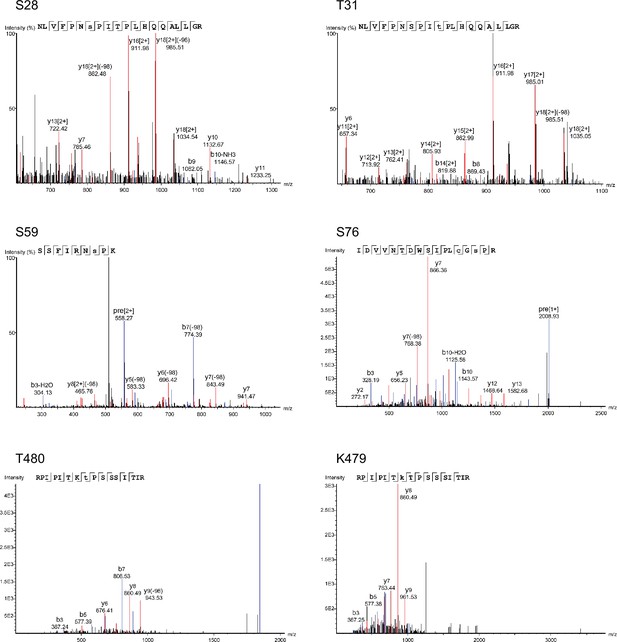
MS spectra from mass spectrometric analyses of Slp1.
Examples of spectra for five phosphorylation sites (S28, T31, S59, S76, and T480) and one ubiquitylation site (K479) identified in Apc15-GFP-assocaited Slp1 purified from metaphase-arrested Padh11-pek1DD cells.
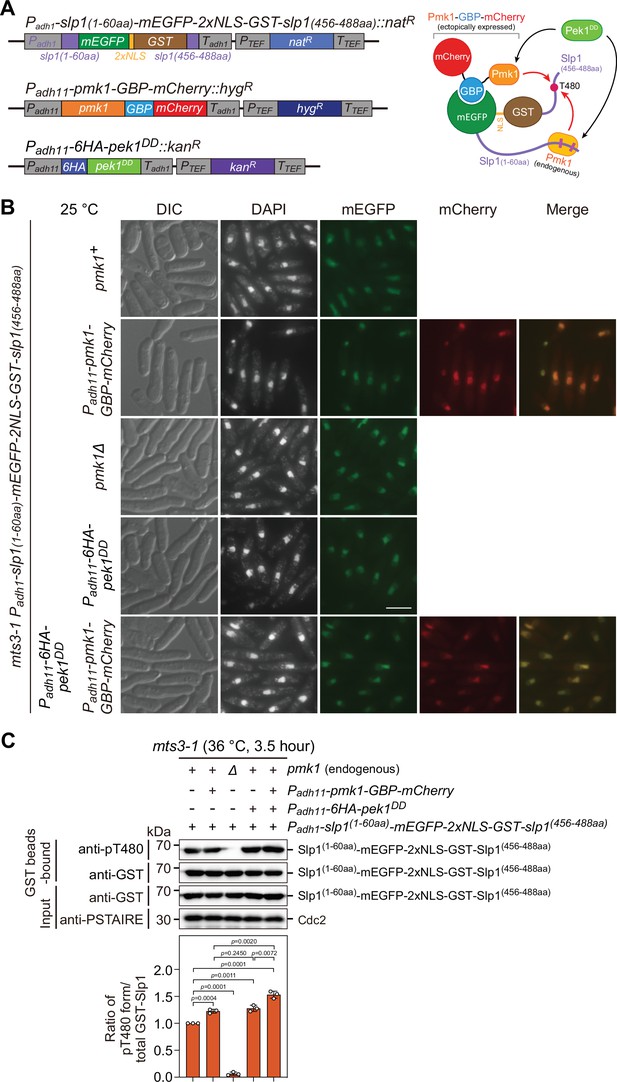
Analysis of Slp1Cdc20 phosphorylation at T480 in vivo upon forced tethering of Slp1Cdc20 to Pmk1.
(A) Schematic depiction of the experiment design for artificially forced tethering of Slp1Cdc20 to Pmk1 using GBP–GFP system. (Left) Structures of the genomically integrated Padh1-slp1(1–60aa)-mEGFP-2xNLS-GST-slp1(456–488aa)::natR at ade6+ locus, Padh11-pmk1-GBP-mCherry::hygR at lys1+ locus, and Padh11-6xHA-pek1DD::kanR at ura4+ locus. (Right) Cartoon depicting the manipulated targeting of mEGFP-2xNLS-GST fusion with Slp1 fragments to GBP-mCherry fusion of Pmk1 mediated by GBP–GFP binding. Note that Thr480 in Slp1 C-terminus could be phosphorylated by either ectopically expressed Pmk1-GBP-mCherry fusion or endogenous Pmk1 activated by Pek1DD. (B) Representative images of mts3-1 cells expressing mEGFP-2xNLS-GST, GBP-mCherry or 6HA fusion proteins driven by promoters Padh1 or Padh11. Cells were grown to early log phase in liquid yeast extract (YE) at 25°C, and then collected, fixed, DAPI-stained, and visualized by using fluorescence microscopy. DIC, differential interference contrast microscopy. Scale bar, 5 μm. (C) Slp1(1–60aa)-mEGFP-2xNLS-GST-Slp1(456–488aa) was purified using GST pull-down from mts3-1 cells with indicated genotypes arrested at 36°C for 3.5 hr, and detected with anti-GST and anti-pThr480 antibodies. Note that pThr480 is absent in pmk1∆ cells, while its phosphorylation level was further enhanced in cells when both Pek1DD and Pmk1-GBP-mCherry were present compared to cells only expressing either fusions. Blots shown are the representative of three independent experiments. pThr480 levels were normalized with ratio between anti-pThr480-recognized and GST bead-bound Slp1 fragment fusion in cells only expressing Slp1(1–60aa)-mEGFP-2xNLS-GST-Slp1(456–488aa) set as 1.0. Error bars indicate mean ± standard deviation of three independent experiments. Two-tailed unpaired t-test was used to derive p values against wild-type cells.
-
Figure 4—figure supplement 3—source data 1
Uncropped blots for Figure 4—figure supplement 3C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp3-data1-v1.pdf
-
Figure 4—figure supplement 3—source data 2
Raw data of quantitative analysis of Slp1 T480 phosphorylation levels for Figure 4—figure supplement 3C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp3-data2-v1.xlsx
-
Figure 4—figure supplement 3—source data 3
Full raw unedited blot (bead-bound, anti-pT480) for Figure 4—figure supplement 3C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp3-data3-v1.zip
-
Figure 4—figure supplement 3—source data 4
Full raw unedited blot (bead-bound, anti-GST) for Figure 4—figure supplement 3C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp3-data4-v1.zip
-
Figure 4—figure supplement 3—source data 5
Full raw unedited blot (input, anti-GST) for Figure 4—figure supplement 3C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp3-data5-v1.zip
-
Figure 4—figure supplement 3—source data 6
Full raw unedited blot (input, Cdc2) for Figure 4—figure supplement 3C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp3-data6-v1.zip
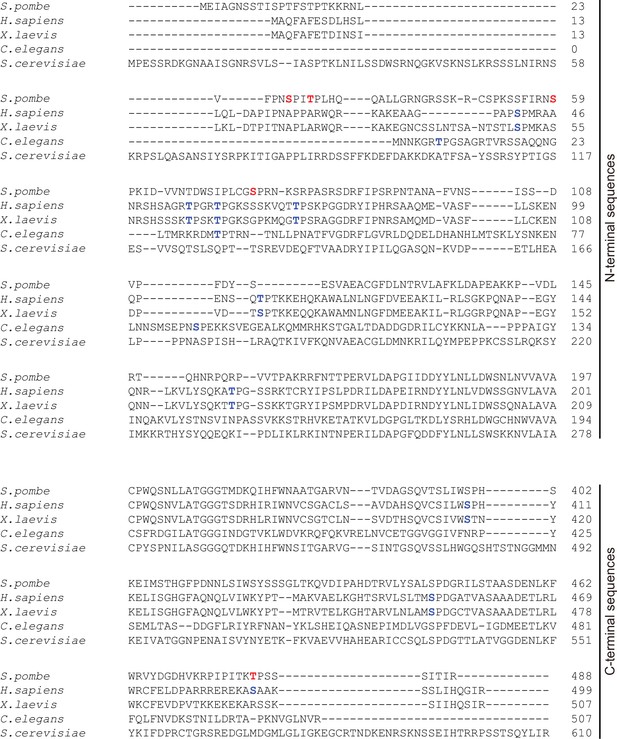
Sequence alignment performed with N- and C-terminal tails of S. pombe Slp1 and its homolog sequences from human (H. sapiens), frog (X. laevis), worm (C. elegans), and budding yeast (S. cerevisiae).
Five phosphorylation sites (Ser28, Thr31, Ser59, Ser76, and Thr480) in Slp1 identified in this study are indicated in red. Putative Cdk1 or mitogen-activated protein kinase (MAPK) phosphorylation sites in Slp1 homologs (e.g. Ser41, Thr55, Thr59, Thr70, Thr106, Thr157, Ser408, Ser452, and Ser487 in human Cdc20; Ser50, Thr64, Thr68, and Thr79 in frog Cdc20; and Thr7, Thr32, and Ser87 in worm Cdc20) suggested by previous studies are indicated in blue.
Mutant proteins of Slp1(T480A) and Slp1(S76A;T480A) are also stabilized in asynchronously growing cells.
Immunoblotting of extracts from asynchronously growing nda3-KM311 cells at 30°C expressing wild-type slp1+ or slp1 with the indicated mutations. An extract from nda3-KM311 slp1+ cells synchronized with HU and arrested at metaphase by treatment at 18°C for 6 hr was loaded as a control. Blots shown are the representative of three independent experiments. Slp1Cdc20 levels were quantified with the relative ratio between Slp1Cdc20 and Cdc2 in wild-type strain asynchronously grown 30°C set as 1.0. Error bars indicate mean ± standard deviation of three independent experiments. Two-tailed unpaired t-test was used to derive p values against wild-type cells.
-
Figure 4—figure supplement 5—source data 1
Uncropped blots for Figure 4—figure supplement 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp5-data1-v1.pdf
-
Figure 4—figure supplement 5—source data 2
Raw data of Slp1 level measurement for Figure 4—figure supplement 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp5-data2-v1.xlsx
-
Figure 4—figure supplement 5—source data 3
Full raw unedited blot (Slp1) for Figure 4—figure supplement 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp5-data3-v1.zip
-
Figure 4—figure supplement 5—source data 4
Full raw unedited blot (Cdc2) for Figure 4—figure supplement 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp5-data4-v1.zip
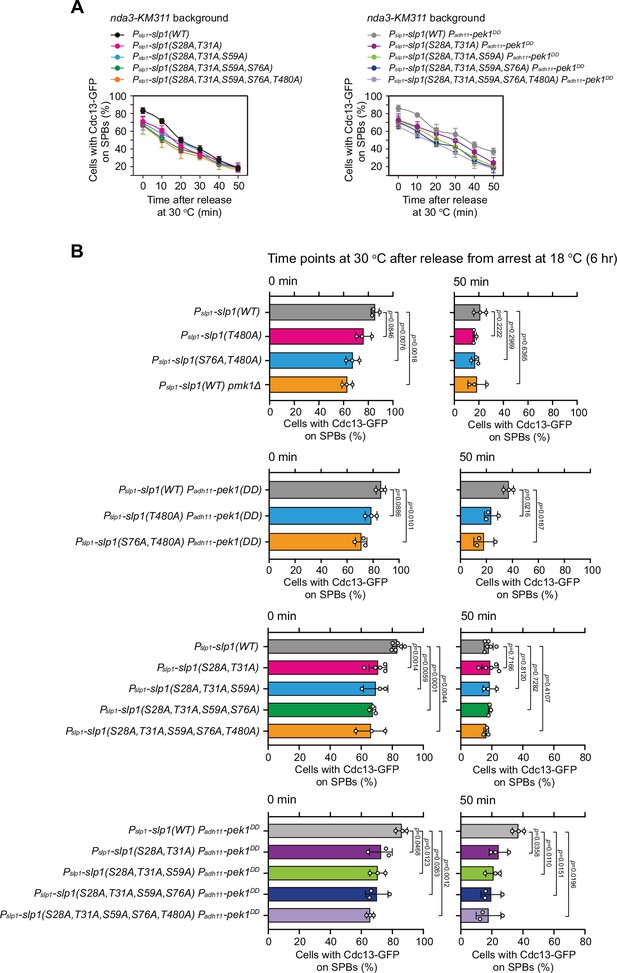
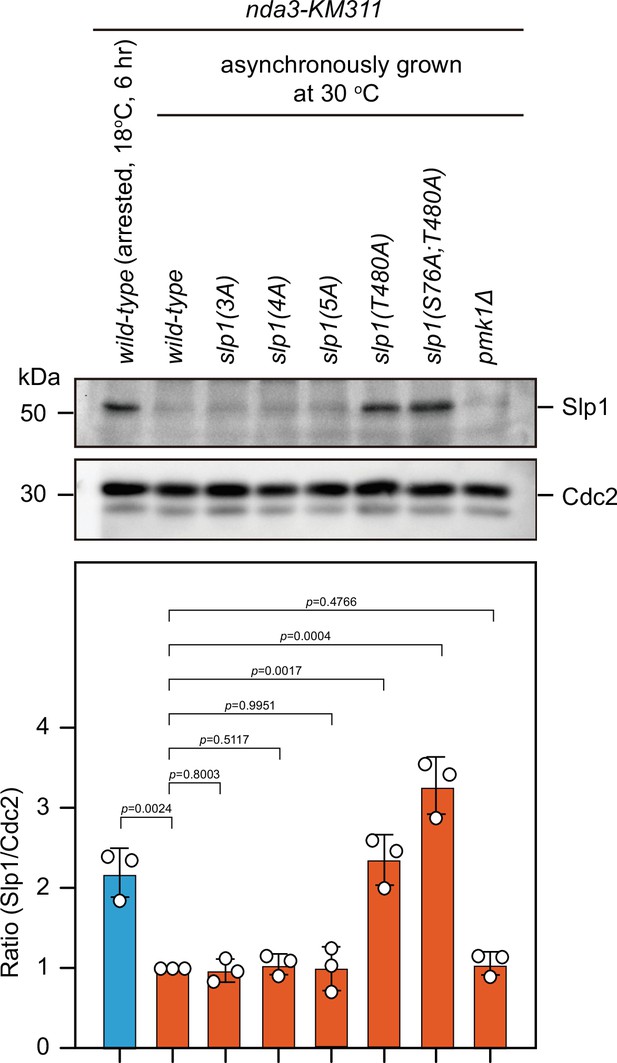
Time-course analyses and quantification of spindle checkpoint inactivation rate in phospho-deficient slp1 mutants at 0 and 50 min after release at 30°C from nda3-mediated arrest.
Cells of indicated strains bearing Cdc13-GFP were grown at 30°C to mid-log phase and arrested at 18°C for 6 hr, and then released at 30°C. For each time point, ≥300 cells were counted for every sample. (A) Time-course analyses of spindle assembly checkpoint (SAC) activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes. Error bars indicate mean ± standard deviation of three independent experiments. (B) Data from time points of 0 and 50 min after release in (A) and Figure 4H were subjected to statistical analysis. Error bars indicate mean ± standard deviation of three independent experiments. p values were calculated against wild-type cells.
-
Figure 4—figure supplement 6—source data 1
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 4—figure supplement 6A and B.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig4-figsupp6-data1-v1.xlsx
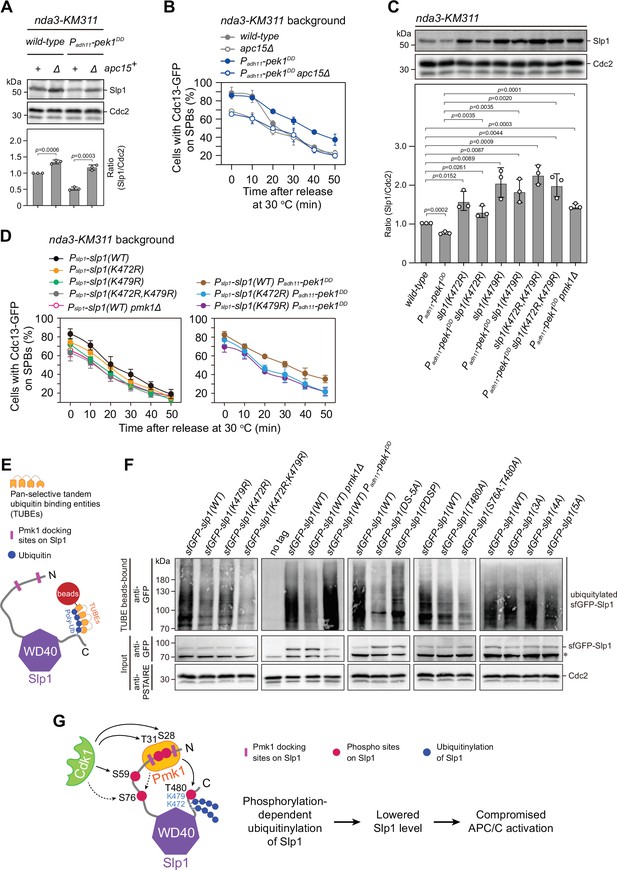
Pmk1- but not Cdk1-mediated Slp1Cdc20 phosphorylation promotes its ubiquitylation.
(A) Immunoblot analysis of Slp1Cdc20 abundance in apc15+or apc15∆ background cells with indicated genotypes after being treated at 18°C for 6 hr. Slp1Cdc20 levels were quantified as in Figure 2B. The experiment was repeated three times. The mean value for each sample was calculated, and p values were calculated against wild-type or pek1DD cells. (B) Time-course analyses of spindle assembly checkpoint (SAC) activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes. The experiments were performed and analyzed as in Figure 1D. (C) Immunoblot analysis of Slp1Cdc20 abundance in K472R, K479R, or K472R/K479R mutants. The experiment was repeated three times. The mean value for each sample was calculated, and p values were calculated against wild-type or pek1DD cells. (D) Time-course analyses of SAC activation and inactivation in nda3-KM311 cdc13-GFP strains with K472R, K479R, or K472R/K479R mutations. The experiments were performed as in (B). (E) Schematic depiction of affinity pull-down assays using TUBE (tandem ubiquitin-binding entity) agarose beads to detect Slp1Cdc20 ubiquitylation. (F) TUBE pull-down assays in mts3-1 strains carrying sfGFP-tagged wild-type or mutants of Slp1Cdc20 or pmk1Δ or pek1DD mutations. The TUBE bead-bound samples were blotted with anti-GFP antibodies. Asterisk indicates unspecific bands recognized by anti-GFP antibodies. (G) Schematic summarizing the negative effect of the coupling of Pmk1 phosphorylation and K472/K479-mediated ubiquitylation on Slp1Cdc20 abundance and anaphase-promoting complex/cyclosome (APC/C) activation.
-
Figure 5—source data 1
Uncropped blots for Figure 5A, C, F.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-data1-v1.pdf
-
Figure 5—source data 2
Raw data of Slp1 level measurement and time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 5A–D.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-data2-v1.xlsx
-
Figure 5—source data 3
Raw blots unedited for Figure 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-data3-v1.zip
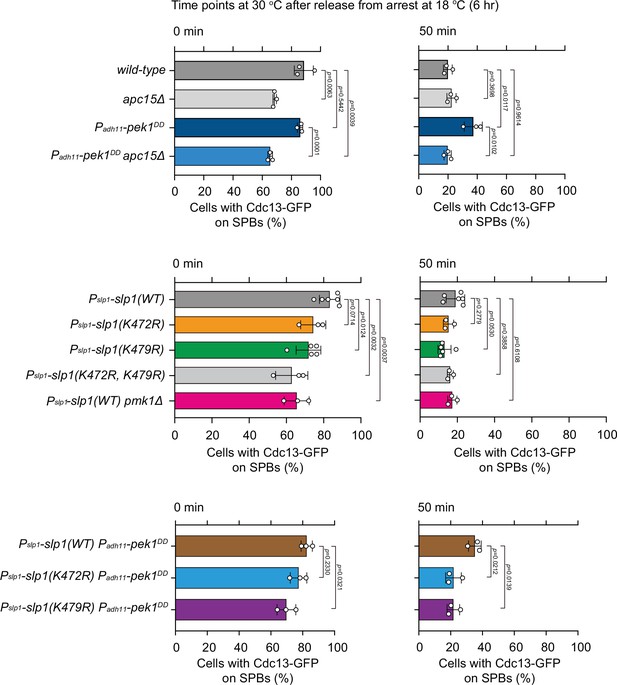
Quantification of spindle checkpoint inactivation rate in ubiquitylation-relevant slp1 mutants at 0 and 50 min after release at 30°C from nda3-mediated arrest.
Cells of indicated strains bearing Cdc13-GFP were grown at 30°C to mid-log phase and arrested at 18°C for 6 hr, and then released at 30°C. For each time point, ≥300 cells were counted for every sample. Data from time points of 0 and 50 min after release in Figure 5B, D were subjected to statistical analysis. Error bars indicate mean ± standard deviation of three independent experiments. p values were calculated against wild-type cells.
-
Figure 5—figure supplement 1—source data 1
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp1-data1-v1.xlsx
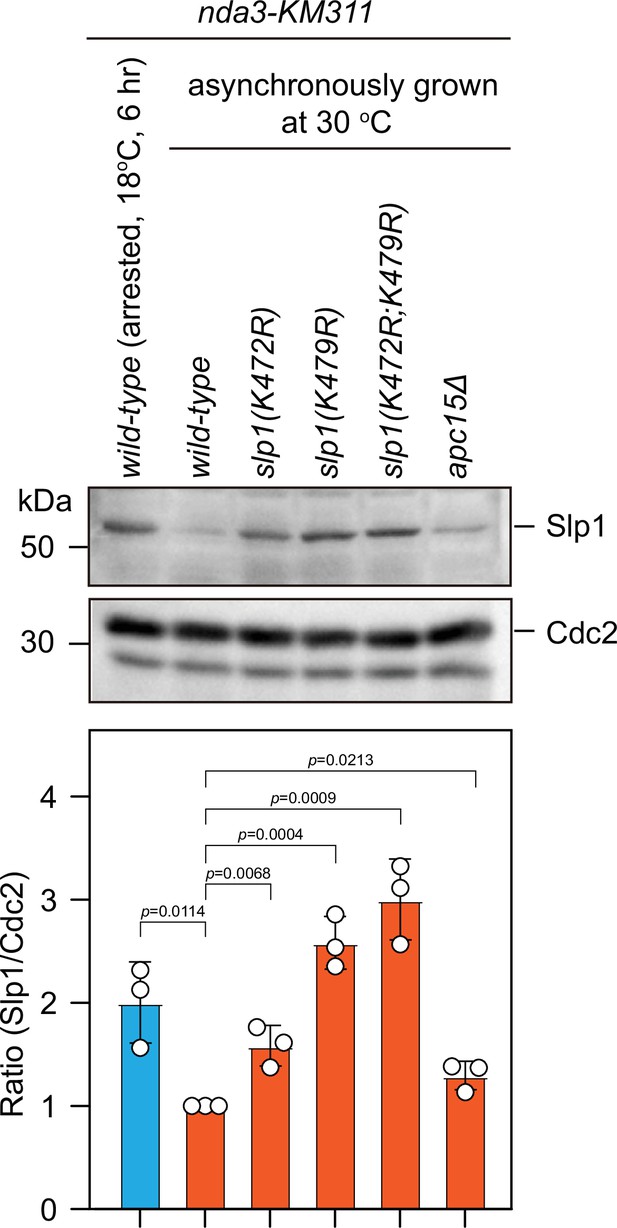
Slp1 proteins with mutations of K479R or K472R are also stabilized in asynchronously growing cells.
Immunoblotting of extracts from asynchronously growing nda3-KM311 cells at 30°C expressing wild-type slp1+ or slp1 with the indicated mutations. A strain with apc15 deletion (apc15Δ) was included as a control for comparison, which has been previously shown to have stabilized Slp1 in interphase cells (Sewart and Hauf, 2017). An extract from nda3-KM311 slp1+ cells synchronized with HU and arrested at metaphase by treatment at 18°C for 6 hr was also loaded as a control. Blots shown are the representative of three independent experiments. Slp1Cdc20 levels were quantified with the relative ratio between Slp1Cdc20 and Cdc2 in wild-type strain asynchronously grown at 30°C set as 1.0. Error bars indicate mean ± standard deviation of three independent experiments. Two-tailed unpaired t-test was used to derive p values against wild-type cells.
-
Figure 5—figure supplement 2—source data 1
Uncropped blots for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp2-data1-v1.pdf
-
Figure 5—figure supplement 2—source data 2
Raw data of Slp1 level measurement for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp2-data2-v1.xlsx
-
Figure 5—figure supplement 2—source data 3
Full raw unedited blot (Slp1) for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp2-data3-v1.zip
-
Figure 5—figure supplement 2—source data 4
Full raw unedited blot (Cdc2) for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp2-data4-v1.zip

Sequence alignment of C-terminal tails of Cdc20 homologs.
Sequence alignment performed with C-terminal tails of S. pombe Slp1 and its Cdc20 homolog sequences from human (H. sapiens), frog (X. laevis), and worm (C. elegans). Lysines residues in fission yeast Slp1 (K472 and K479) and human Cdc20 (K485 and K490), which have been confirmed in this study and previous studies (Danielsen et al., 2011; Mansfeld et al., 2011), respectively, to be responsible for their ubiquitylation and degradation, are indicated in red. Other lysine residues and flanking threonine or serine residues in these sequences are also highlighted in orange or green, respectively.
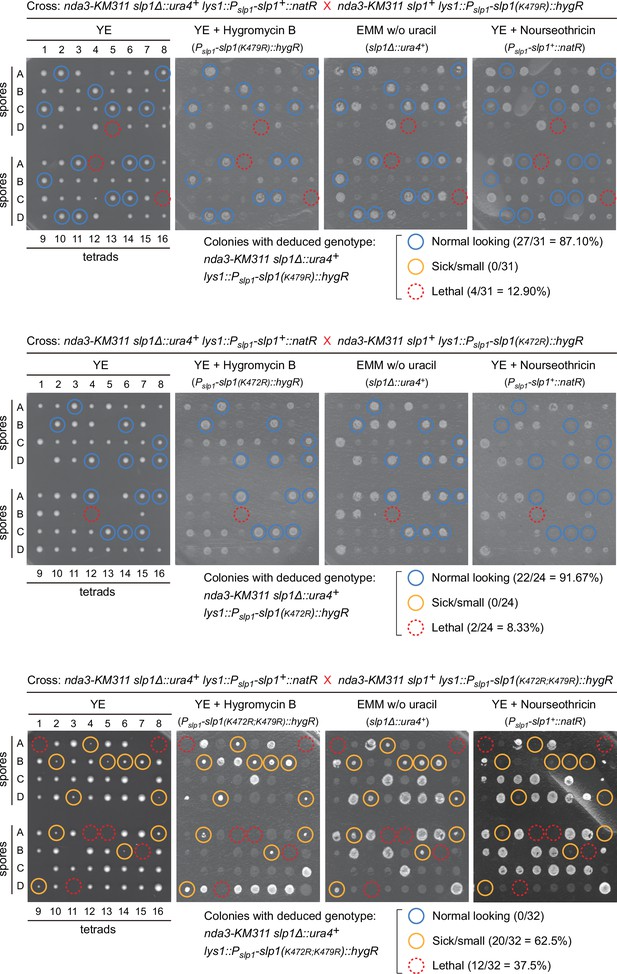
Viability of slp1(K472R;K479R) cells is compromised.
Normal-looking four-spore asci obtained after crosses between nda3-KM311 background strains carrying slp1+ lys1::Pslp1-slp1(K472R)::hygR, slp1+ lys1::Pslp1-slp1(K479R)::hygR or slp1+ lys1::Pslp1-slp1(K472R;K479R)::hygR and slp1Δ::ura4+ lys1::Pslp1-slp1+::natR strain were dissected using a micromanipulator. The genotypes of colonies formed from germinated spores were deduced after being replicated on selective plates. Mutant candidates of nda3-KM311 slp1Δ::ura4+ lys1::Pslp1-slp1(K472R)/(K479R)/(K472R;K479R)::hygR are indicated by light blue circles (normal-looking colonies), yellow circles (small colonies), or dashed red circles (spores failing to germinate) on representative plates. Quantitative analyses of synthetic lethality of desired mutants based on dissected four-spore asci showed that simultaneous mutation of K472 and K479 in Slp1 to arginine (i.e. K472R;K479R double mutant) causes strong synthetic growth defects.
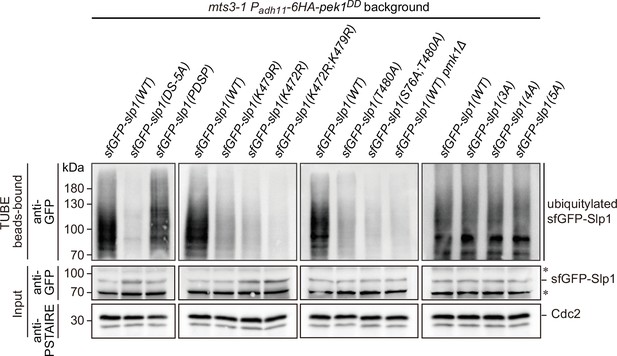
Enhanced Slp1Cdc20 ubiquitylation in pek1DD cells can be removed by Pmk1-docking-, Pmk1-phosphorylation- or ubiquitylation-deficient mutations in Slp1Cdc20.
Cultures of mts3-1 his5::Padh11-pek1DD::natR strains carrying sfGFP-tagged wild-type or mutants of Slp1Cdc20 with mutations in Pmk1-docking motives (DS-5A, PDSP), ubiquitylation sites (K472R; K479R or K472R/K479R), Pmk1 phosphorylation sites (T480A; S76A/T480A), or combined Pmk1 and Cdk1 phosphorylation sites (3A, 4A, or 5A) were first grown at 25°C to mid-log phase and then shifted to 36°C for 3.5 hr to block cells in mitosis prior to harvesting and cell lysis. The strain mts3-1 his5::Padh11-pek1DD::natR pmk1Δ served as a negative control. Ubiquitinated proteins were pulled down from yeast lysates using tandem ubiquitin-binding entities (TUBEs). The TUBE bead-bound samples were blotted with anti-GFP antibodies. Asterisks indicate unspecific bands recognized by anti-GFP antibodies.
-
Figure 5—figure supplement 5—source data 1
Uncropped blots for Figure 5—figure supplement 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp5-data1-v1.pdf
-
Figure 5—figure supplement 5—source data 2
Full raw unedited blot (bead-bound sfGFP-Slp1, blot 1) for Figure 5—figure supplement 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp5-data2-v1.zip
-
Figure 5—figure supplement 5—source data 3
Raw unedited blots for Figure 5—figure supplement 5.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig5-figsupp5-data3-v1.zip
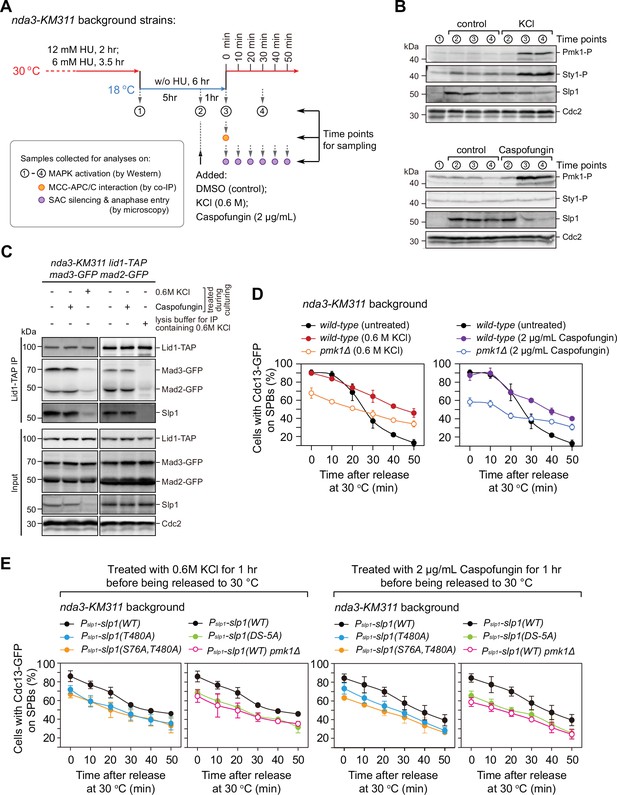
Osmotic stress and cell wall damage trigger rapid Pmk1 phosphorylation-dependent Slp1Cdc20 downregulation and mitotic exit delay.
(A) Schematic depiction of the experimental design for treatment with KCl or caspofungin during nda3-mediated spindle checkpoint activation to activate mitogen-activated protein kinases (MAPKs). Samples were collected at indicated time points for subsequent analyses including immunoblotting, co-immunoprecipitation (co-IP), and time-course analysis on spindle assembly checkpoint (SAC) or anaphase-promoting complex/cyclosome (APC/C) activation. (B) Immunoblot analysis of activation of MAPKs and Slp1Cdc20 protein levels. Samples with or without indicated treatments were blotted with anti-phospho p42/44 and anti-phospho p38 antibodies as indicative of phosphorylated Pmk1 (Pmk1-P) or phosphorylated Sty1 (Sty1-P), respectively. Slp1Cdc20 levels were detected with anti-Slp1 antibodies and anti-Cdc2 was used as loading control. (C) Co-immunoprecipitation analysis of APC/C–mitotic checkpoint complex (MCC) association upon environmental stress. Lid1-TAP was immunoprecipitated from nda3-KM311-arrested cells and associated Mad2-GFP, Mad3-GFP and Slp1Cdc20 were detected as in Figure 2A. Note that APC/C–MCC association was disrupted when 0.6 M KCl was present during cell culturing or during immunoprecipitation procedures. (D) Time-course analyses of SAC activation and inactivation in nda3-KM311 cdc13-GFP strains with indicated genotypes after arrest at 18°C and KCl or caspofungin treatments. The experiment was repeated three times and the mean value and p value for each sample were calculated as in Figure 1D. (E) Time-course analyses of SAC activation and inactivation efficiency in Pmk1-docking- and phosphorylation-deficient slp1 mutants under environmental stresses elicited by 0.6 M KCl or 2 μg/ml caspofungin.
-
Figure 6—source data 1
Uncropped blots for Figure 6B, C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-data1-v1.pdf
-
Figure 6—source data 2
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 6D, E.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-data2-v1.xlsx
-
Figure 6—source data 3
Raw unedited blots for Figure 6.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-data3-v1.zip
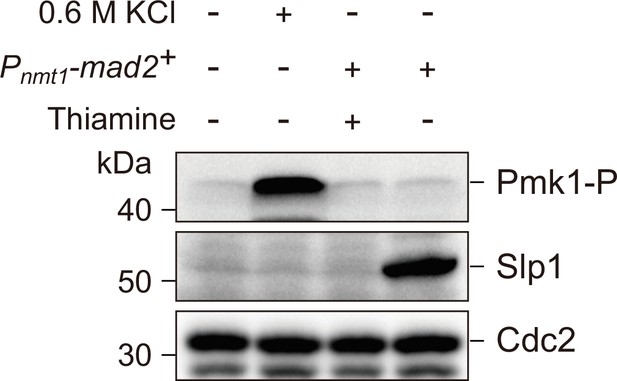
Activation of spindle assembly checkpoint (SAC) by Mad2 overexpression does not trigger Pmk1 activation.
Yeast strain carrying Pnmt1-mad2::leu1+ was pre-cultured in minimal medium with supplements (EMM5S) and 15 µM thiamine. Cells were then washed in EMM5S to remove thiamine before being grown in EMM5S at 30°C for 18 hr to induce Mad2 overexpression. Wild-type cells grown in yeast extract (YE) were treated with 0.6 M KCl for 60 min as a positive control for Pmk1 activation. Protein samples after SDS–PAGE were blotted with anti-phospho p42/44 antibodies as indicative of phosphorylated Pmk1 (Pmk1-P). Slp1Cdc20 levels were detected with anti-Slp1 antibodies and anti-PSTAIRE (detecting Cdc2) was used as loading control. Blots shown are the representative of three independent experiments.
-
Figure 6—figure supplement 1—source data 1
Uncropped blots for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp1-data1-v1.pdf
-
Figure 6—figure supplement 1—source data 2
Full raw unedited blot (phosphorylated Pmk1) for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp1-data2-v1.zip
-
Figure 6—figure supplement 1—source data 3
Full raw unedited blot (Slp1) for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp1-data3-v1.zip
-
Figure 6—figure supplement 1—source data 4
Full raw unedited blot (Cdc2) for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp1-data4-v1.zip
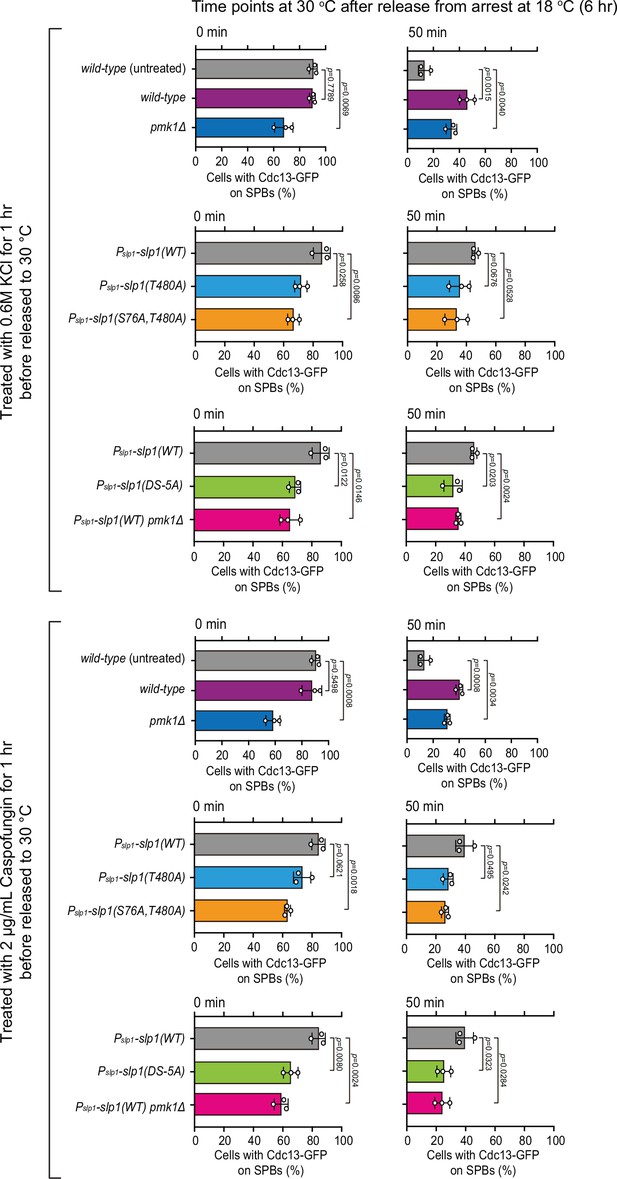
Quantification of spindle checkpoint inactivation rate in the presence of KCl or caspofungin treatment at 0 and 50 min after release at 30°C from nda3-mediated arrest.
Cells of indicated strains bearing Cdc13-GFP were grown at 30°C to mid-log phase and arrested at 18°C for 6 hr, and then released at 30°C. For each time point, ≥300 cells were counted for every sample. Data from time points of 0 and 50 min after release in Figure 6D, E were subjected to statistical analysis. Error bars indicate mean ± standard deviation of three independent experiments. p values were calculated against wild-type cells.
-
Figure 6—figure supplement 2—source data 1
Raw data of time-course analyses of Cdc13-GFP at spindle pole body (SPB) for Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp2-data1-v1.xlsx
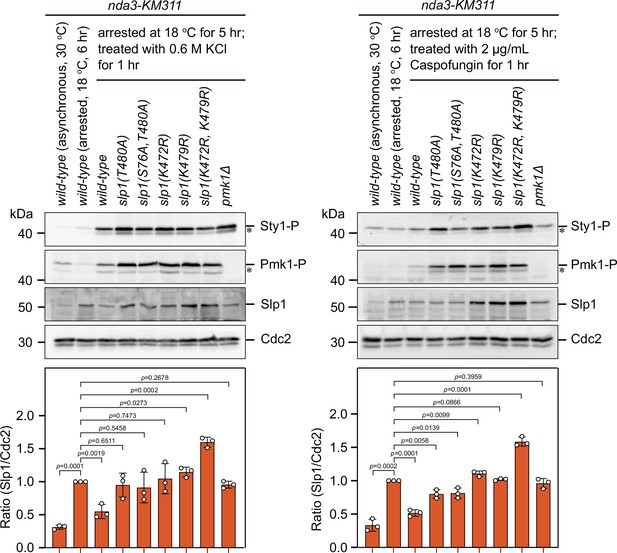
Immunoblot analysis of activation of mitogen-activated protein kinases (MAPKs) and Slp1Cdc20 protein levels in Pmk1 phosphorylation- and ubiquitylation-deficient slp1 mutants under stress.
nda3-KM311 strains expressing wild-type slp1+ or slp1 with the indicated mutations were first synchronized with HU and then arrested at metaphase by treatment at 18°C for 5 hr. Then, a final concentration of 0.6 M KCl or 2 μg/ml caspofungin was added into cultures and left at 18°C for 1 hr before harvesting. nda3-KM311 slp1+ cells either grown asynchronously at 30°C or synchronized with HU and arrested at metaphase by treatment at 18°C for 6 hr served as controls. Protein samples were subjected to SDS–PAGE and immunoblotted with anti-phospho p42/44 and anti-phospho p38 antibodies as indicative of phosphorylated Pmk1 (Pmk1-P) or phosphorylated Sty1 (Sty1-P), respectively. Slp1Cdc20 levels were detected with anti-Slp1 antibodies and anti-PSTAIRE (detecting Cdc2) was used as loading control. The experiment was repeated three times. Slp1Cdc20 levels were quantified with the relative ratio between Slp1Cdc20 and Cdc2 in wild-type strain synchronized and arrested at 18°C for 6 hr set as 1.0. The mean value for each sample was calculated from three independent experiments, and p values were calculated against wild-type cells synchronized and arrested at 18°C for 6 hr. Asterisks indicate unspecific or degraded protein bands.
-
Figure 6—figure supplement 3—source data 1
Uncropped blots for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp3-data1-v1.pdf
-
Figure 6—figure supplement 3—source data 2
Raw data of Slp1 level measurement for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp3-data2-v1.xlsx
-
Figure 6—figure supplement 3—source data 3
Raw unedited blots for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig6-figsupp3-data3-v1.zip
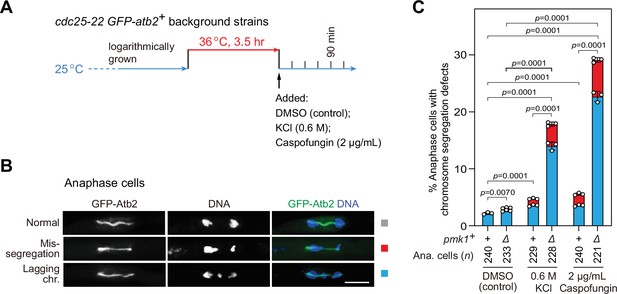
pmk1Δ cells are defective in faithful chromosome segregation upon environmental stress.
(A) Schematic depiction of the experiment design for arrest-and-release of cdc25-22 GFP-atb2+ strains. Environmental stresses were imposed by 0.6 M KCl or 2 μg/ml caspofungin. Samples were taken at 90 min after being shifted back from 36 to 25°C to enrich anaphase cells. (B, C) Analyses of chromosome segregation in anaphase cells treated without or with KCl or caspofungin and after being fixed and stained with DAPI. Example pictures of anaphase cells with long spindles judged by GFP-Atb2 signals are shown (B). Anaphase cells with two categories of chromosome segregation defects (i.e. unequal chromosome segregation (mis-segregation) and lagging chromosomes) were quantified (C). >200 cells were counted for every sample. The mean values for each category were calculated, error bars indicate mean ± standard deviation of three independent experiments. p values were calculated with pooled data of two categories for each sample. n, numbers of anaphase cells analyzed. Scale bar, 5 μm.
-
Figure 7—source data 1
Raw data of defective chromosome segregation analyses for Figure 7C.
- https://cdn.elifesciences.org/articles/97896/elife-97896-fig7-data1-v1.xlsx
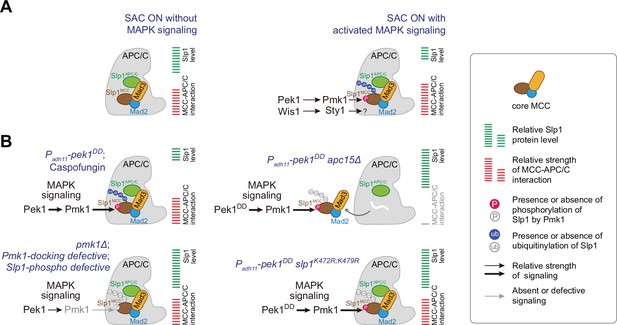
Summary of the mechanisms that how mitogen-activated protein kinases (MAPKs) negatively regulate APC/C activity in fission yeast.
(A) Schematic depiction of a possible dual mechanism that how activated MAPK signaling pathways are involved in delaying APC/C activation and spindle assembly checkpoint (SAC) inactivation. Upon SAC and MAPK signaling activation, division of labor between the cell integrity pathway (CIP) and the stress-activated pathway (SAP) enables the phosphorylation of Slp1Cdc20 and unidentified substrate(s) by Pmk1 and Sty1, respectively, which leads to lowered Slp1Cdc20 level and enhanced mitotic checkpoint complex (MCC) affinity for APC/C. (B) Summary of the alteration of Slp1Cdc20 protein levels and MCC–APC/C association strength under various conditions examined in this study. White wavy line indicates the absence of Apc15.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/97896/elife-97896-mdarchecklist1-v1.doc
-
Supplementary file 1
Yeast strains used in this study.
- https://cdn.elifesciences.org/articles/97896/elife-97896-supp1-v1.doc





