A genome-wide association study implicates the olfactory system in Drosophila melanogaster diapause-associated lifespan extension and fecundity
Figures
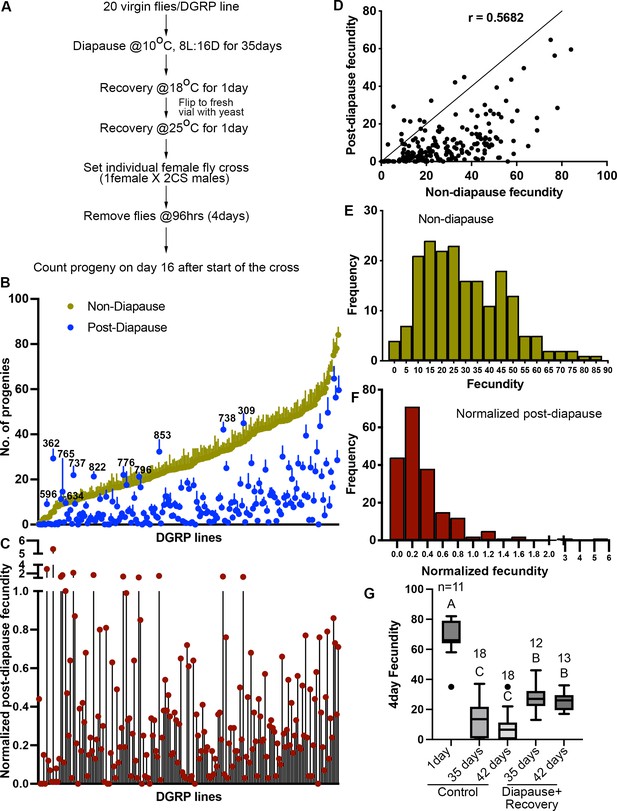
Quantification of diapause in Drosophila Genetic Reference Panel (DGRP) lines measured as the ratio of post-diapause to non-diapause fecundity.
(A) Schematic of experimental workflow. (B) Average number of progenies produced (fecundity) in a 4-day individual female fly mating experiment of DGRP lines either as non-diapausing (yellow) or after a 35-day post-diapause (blue) virgin flies. Each dot represents the average fecundity, and the line above represents the standard error. DGRP line numbers are indicated wherever the post-diapause fecundity exceeds the non-diapause fecundity. (C) Normalized post-diapause fecundity average of (individual post-diapause fecundity/mean non-diapause fecundity) of each DGRP line. (D) Correlation of post-diapause to non-diapause fecundity. Pearson’s r=0.5682. r2=0.3228. (E–F) Frequency distribution of DGRP lines fecundity under non-diapause (E) and of the normalized post-diapause fecundity (F). (G) Average 4-day fecundity of single female flies, each crossed with two young Canton-S male flies, aged for 1, 35, or 42 days in non-diapause conditions or kept in diapause conditions for 35 or 42 days followed by recovery. One-way ANOVA and Tukey’s multiple comparison test, compact letter display shows comparisons. n is the number of individual female fly fecundity measured, and whiskers represent the smallest and largest values within 1.5× the interquartile range (IQR).
-
Figure 1—source data 1
Non-diapause and post-diapause fecundity in Drosophila Genetic Reference Panel (DGRP) lines.
Quantification of fecundity of DGRP lines in non-diapausing and post-diapausing conditions.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Excel sheet containing data corresponding to Figure 1D–F.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Fecundity of Canton-S control flies scored at different ages and conditions.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig1-data3-v1.xlsx
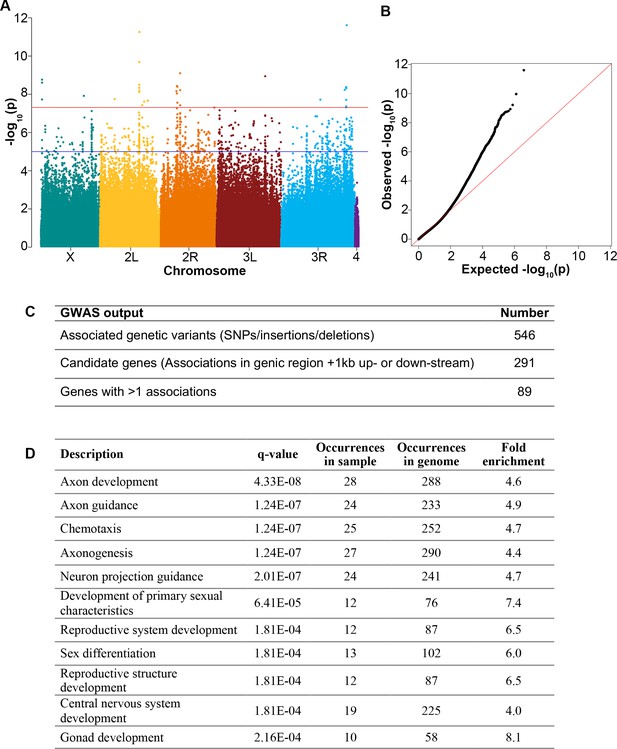
Genome-wide association of Drosophila diapause.
(A) Manhattan plot for genome-wide association distribution. The position of each point along the y-axis indicates –log10(p-value) of association of a single nucleotide polymorphism (SNP), insertion, or deletion. Points above the blue line have a p-value<1e–5. The red line represents Bonferroni-corrected p-value = 4.8e–8. (B) Q-Q plot of p-values from the Drosophila Genetic Reference Panel (DGRP) single variant genome-wide association study (GWAS) with the red line representing expected p-value and observed p-values deviating (black dots) from expected. (C) Numbers of genetic variants and candidate genes associated with diapause according to the GWAS. (D) Subnetworks from the Cytoscape analysis showing q-value (using the Benjamini-Hochberg procedure) for each subnetwork identified.
-
Figure 2—source data 1
Gene variants associated with post-diapause fecundity identified by genome-wide association study (GWAS).
Results using the DGRP2 tool to analyze the ratio of post-diapause to non-diapause fecundity.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Candidate diapause-associated genes identified by genome-wide association study (GWAS).
Genes within 1 kb upstream or downstream of the gene variants associated with diapause are candidate diapause-associated genes. The number of different associations is provided along with references for genes previously associated with diapause.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig2-data2-v1.xlsx
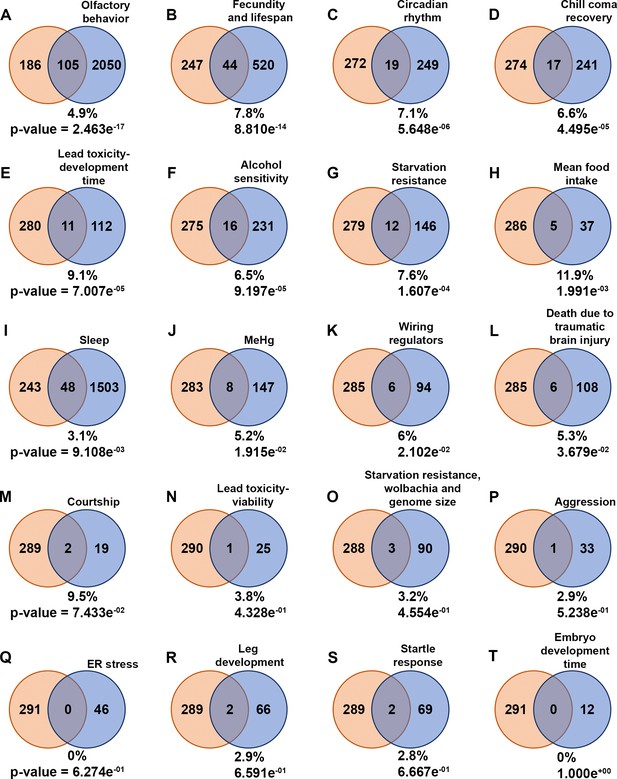
Common genes to diapause-genome-wide association study (GWAS) hits and other behavior-associated genes.
(A–T) Venn diagrams illustrate the intersection of genes associated with diapause identified through GWAS (diapause-GWAS) with genes from other behavior-related gene lists obtained from various studies. The diapause-GWAS gene set is represented as the first set throughout the figure, while subsequent sets represent different behavior-related gene lists identified in separate studies. The percentage of common genes compared to the total genes from different respective behavior-associated gene lists is provided for each Venn diagram. p-Values of overlap to the diapause gene list determined by Fisher’s exact tests are also provided. Venn diagrams are arranged in the order of p-values.
-
Figure 3—source data 1
Comparison of diapause-genome-wide association study (GWAS) gene list to other behavior-associated gene lists.
Diapause-GWAS gene lists are compared to other behavior-associated gene lists from different studies (DOIs are included in each tab from where gene lists are obtained). Common genes identified, Venn diagram, and p-values are added in each tab of the Excel sheet.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig3-data1-v1.xlsx
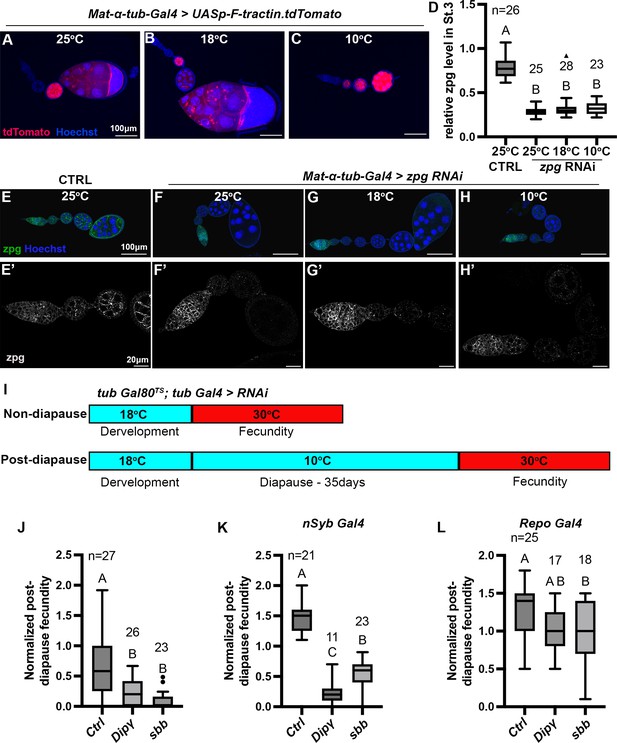
RNAi-mediated loss-of-function study to identify genes involved in diapause.
(A–C) Mat-ɑ-tub-Gal4 driving expression of UASp-F-tractin.tdTomato (red) at the indicated temperatures. Scale bars are 100 µm. (D) Quantification of zpg RNAi knockdown in stage 3 egg chambers normalized to the level of Zpg in the germarium (one-way ANOVA and Tukey’s multiple comparison test, compact letter display shows comparisons). The numbers (n) of stage 3 egg chambers quantified are shown, and whiskers represent the smallest and largest values within 1.5× the interquartile range (IQR). (E–H) Representative images of egg chambers stained with anti-Zpg antibody (green) from either control (no-knockdown) (E, E’) or knockdown of zpg (F-H’) driven by Mat-ɑ-tub-Gal4 at different temperatures. (E’–H’) are higher magnification, single channel views of the ovarioles shown in (E–H). Scale bars are 100 µm in (E–H) and 20 µm in (E’–H’). All flies in (A–H’) were kept at respective temperatures for 3 weeks. (I) Experimental design for RNAi knockdown specifically during recovery for the experiment shown in (J). The temperature-sensitive Gal80 repressor of Gal4 prevented RNAi expression during development and diapause. Incubation at 30°C during recovery inactivates Gal80, allowing Gal4-mediated RNAi knockdown. (J) Ubiquitous knockdown of Dip-γ or sbb with tub Gal4 specifically during recovery as shown in (I) significantly reduces post-diapause/non-diapause fecundity compared to the control (tubGal80TS;tubGal4 >Ctrl RNAi #9331). (K) Pan-neuronal RNAi knockdown of Dip-γ and sbb with nSybGal4 significantly reduces post-diapause/non-diapause fecundity compared to the control (nSyb Gal4>Ctrl RNAi #54037). (L) Glia-specific knockdown of Dip-γ or sbb with Repo Gal4 causes little or no reduction in post-diapause/non-diapause fecundity (Control- Repo Gal4>Ctrl RNAi #54037). In (J–L), one-way ANOVA and Tukey’s multiple comparison test, compact letter display shows comparisons. n is the number of individual female flies tested, and whiskers represent the smallest and largest values within 1.5× the interquartile range (IQR).
-
Figure 4—source data 1
Excel sheet containing data corresponding to Figure 4D.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig4-data1-v1.xlsx
-
Figure 4—source data 2
RNAi screen of top 15 genome-wide association study (GWAS)-associated genes.
Fecundity assessment of RNAi-mediated knockdown of different genes from the GWAS list. Non-diapause and post-diapause fecundity were measured, and post-diapause fecundity was normalized by dividing individual post-diapause female fly fecundity by average non-diapause fecundity.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Tissue-specific RNAi of Dip-γ and sbb.
Fecundity assessment of pan-neuronal and glial-specific RNAi-mediated knockdown of Dip-γ and sbb.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig4-data3-v1.xlsx
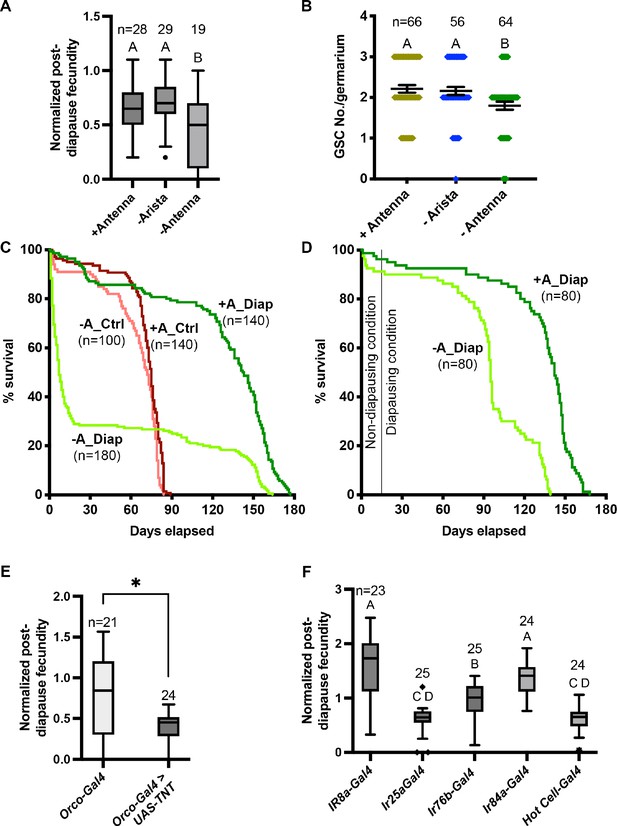
Neural control of diapause.
(A) Effect of antenna removal on recovery of fecundity post-diapause. Arista removal was used as a control for the surgery. n is the number of individual female flies tested. One-way ANOVA and Tukey’s multiple comparison test, compact letter display shows comparisons. Whiskers represent the smallest and largest values within 1.5× the interquartile range (IQR). (B) Effect of antenna removal on germline stem cell (GSC) recovery after 5 weeks of diapause. n is the number of germaria counted (there are typically 2–3 GSCs/germarium). One-way ANOVA and Tukey’s multiple comparison test with compact letter display to show comparisons. Error bars represent standard error. (C) Role of antenna in lifespan extension in diapause. Control flies were maintained at 25°C and diapause flies were moved to 10°C. Median survival for flies with intact antenna in diapause (+A_Diap) – 142.5 days; antennaless flies in diapause (-A_Diap) – 7 days; control with intact antenna in optimal conditions (+A_Ctrl) – 75 days; and antennaless flies in optimal conditions (-A_Ctrl) – 72 days. Survival curves are compared pairwise using the Log-rank (Mantel-Cox) test and Χ2 values are: Diap±antenna = 98.89, Ctrl ±antenna = 11.69, Diap+antenna vs Ctrl+antenna = 163.5, and Diap – antenna vs Ctrl – antenna=0.5. n represents the number of flies used for the survival curve. (D) Control (+A_Diap) and antennaless (-A_Diap) flies were maintained at 25ºC for 2 weeks post-surgery to allow for wound healing and shifted to the diapausing conditions. Median survival for +A_Diap – 142 days; -A_Diap – 95 days. Survival curves are compared pairwise using the Log-rank (Mantel-Cox) test and Χ2 values = 86. n represents the number of flies used for the survival curve. (E–F) Effects on diapause of inactivating neuronal transmission by driving tetanus toxin using UAS-TeTxLC.tdt, (E) in odor responsive neurons using the indicated Gal4 lines. Orco is a co-receptor for odorant receptors of the OR class. (F) Ir8a is a co-receptor involved in organic acid detection. Ir25a is a co-receptor involved in chemo- and thermo-sensation. Ir76b is involved in the detection of various amines and salt. Ir84a is involved in the detection of phenylacetic acid and male courtship behavior. Hot cells are heat-sensitive cells in the arista. In (E-F), one-way ANOVA and Tukey’s multiple comparison test, compact letter display shows comparisons. n is the number of individual female flies tested, and whiskers represent the smallest and largest values within 1.5× the interquartile range (IQR).
-
Figure 5—source data 1
Role of antennae in the ability of flies to undergo diapause.
Fecundity assay of Canton-S control flies with antennae or aristae removed, along with controls.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Excel sheet containing data corresponding to Figure 5B–D.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig5-data2-v1.xlsx
-
Figure 5—source data 3
Identification of specific cell types involved in diapause in fly antennae.
Fecundity assay used to identify diapause-modifying cell types in fly antennae by specifically blocking neuronal transmission through the expression of tetanus toxin.
- https://cdn.elifesciences.org/articles/98142/elife-98142-fig5-data3-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | DGRP lines | BDSC; Huang et al., 2014 | RRID:SCR_006457 | |
| Genetic reagent (D. melanogaster) | 2704 | BDSC | RRID:BDSC_27049 | |
| Genetic reagent (D. melanogaster) | 32034 | BDSC | RRID:BDSC_32034 | |
| Genetic reagent (D. melanogaster) | 40889 | BDSC | RRID:BDSC_40889 | |
| Genetic reagent (D. melanogaster) | 27508 | BDSC | RRID:BDSC_27508 | |
| Genetic reagent (D. melanogaster) | 54811 | BDSC | RRID:BDSC_54811 | |
| Genetic reagent (D. melanogaster) | 44661 | BDSC | RRID:BDSC_44661 | |
| Genetic reagent (D. melanogaster) | 53315 | BDSC | RRID:BDSC_53315 | |
| Genetic reagent (D. melanogaster) | 54037 | BDSC | RRID:BDSC_54037 | |
| Genetic reagent (D. melanogaster) | 57840 | BDSC | RRID:BDSC_57840 | |
| Genetic reagent (D. melanogaster) | 63013 | BDSC | RRID:BDSC_63013 | |
| Genetic reagent (D. melanogaster) | 56867 | BDSC | RRID:BDSC_56867 | |
| Genetic reagent (D. melanogaster) | 80461 | BDSC | RRID:BDSC_80461 | |
| Genetic reagent (D. melanogaster) | 35785 | BDSC | RRID:BDSC_35785 | |
| Genetic reagent (D. melanogaster) | 9331 | BDSC | RRID:BDSC_9331 | |
| Genetic reagent (D. melanogaster) | 5138 | BDSC | RRID:BDSC_5138 | |
| Genetic reagent (D. melanogaster) | 7019 | BDSC | RRID:BDSC_7019 | |
| Genetic reagent (D. melanogaster) | 23292 | BDSC | RRID:BDSC_23292 | |
| Genetic reagent (D. melanogaster) | 41731 | BDSC | RRID:BDSC_41731 | |
| Genetic reagent (D. melanogaster) | 41728 | BDSC | RRID:BDSC_41728 | |
| Genetic reagent (D. melanogaster) | 51311 | BDSC | RRID:BDSC_51311 | |
| Genetic reagent (D. melanogaster) | 41750 | BDSC | RRID:BDSC_41750 | |
| Genetic reagent (D. melanogaster) | 28838 | BDSC | RRID:BDSC_28838 | |
| Genetic reagent (D. melanogaster) | 35607 | BDSC | RRID:BDSC_35607 | |
| Genetic reagent (D. melanogaster) | 7063 | BDSC | RRID:BDSC_7063 | |
| Genetic reagent (D. melanogaster) | 58989 | BDSC | RRID:BDSC_58989 | |
| Genetic reagent (D. melanogaster) | 51635 | BDSC | RRID:BDSC_51635 | |
| Genetic reagent (D. melanogaster) | 64349 | BDSC | RRID:BDSC_64349 | |
| Genetic reagent (D. melanogaster) | 7415 | BDSC | RRID:BDSC_7415 | |
| Genetic reagent (D. melanogaster) | 44077 | VDRC | RRID:Flybase_FBst046539 | |
| Genetic reagent (D. melanogaster) | 330386 | VDRC | RRID:Flybase_FBst0491275 | |
| Genetic reagent (D. melanogaster) | 10268 | VDRC | RRID:Flybase_FBst0450061 | |
| Genetic reagent (D. melanogaster) | 40698 | VDRC | RRID:Flybase_FBst0463709 | |
| Genetic reagent (D. melanogaster) | 991 | VDRC | RRID:Flybase_FBst0471623 | |
| Genetic reagent (D. melanogaster) | 992 | VDRC | RRID:Flybase_FBst0471628 | |
| Genetic reagent (D. melanogaster) | 21052 | VDRC | RRID:Flybase_FBst0453901 | |
| Genetic reagent (D. melanogaster) | 100412 | VDRC | RRID:Flybase_FBst0472285 | |
| Genetic reagent (D. melanogaster) | 51936 | VDRC | RRID:Flybase_FBst0469629 | |
| Genetic reagent (D. melanogaster) | 106911 | VDRC | RRID:Flybase_FBst0478734 | |
| Genetic reagent (D. melanogaster) | 104551 | VDRC | RRID:Flybase_FBst0478734 | |
| Genetic reagent (D. melanogaster) | 17767 | VDRC | RRID:Flybase_FBst0476409 | |
| Genetic reagent (D. melanogaster) | 104056 | VDRC | RRID:Flybase_FBst0452858 | |
| Genetic reagent (D. melanogaster) | 60102 | VDRC | RRID:Flybase_FBst0475914 | |
| Genetic reagent (D. melanogaster) | 013805 | VDRC | RRID:SCR_013805 | |
| Genetic reagent (D. melanogaster) | 101659 | VDRC | RRID:Flybase_FBst0473532 | |
| Genetic reagent (D. melanogaster) | 36166 | VDRC | RRID:Flybase_FBst0461553 | |
| Genetic reagent (D. melanogaster) | Hot cell-Gal4 | Zuker lab, Gallio et al., 2011 | DOI: 10.1016 /j.cell.2011.01.028 | |
| Genetic reagent (D. melanogaster) | Ir21a-Gal4 | Craig Montell lab, Ni et al., 2016 | DOI: https://doi.org/10.7554/eLife.13254 | |
| Antibody | anti-Zpg antibody (rabbit polyclonal) | Smendziuk et al., 2015 | DOI: https://doi.org/10.1242/dev.123448 | 1:20,000 |
| Antibody | anti-Hts antibody (1B1) (mouse monoclonal) | DSHB | RRID:AB_528070 | 1:20 |
| Antibody | anti-Vasa antibody (rat monoclonal) | DSHB | RRID:AB_760351 | 1:20 |
| Software, algorithm | Prism | GraphPad Version 10.4.2 (534) | RRID:SCR_002798 | |
| Software, algorithm | Cytoscape | Shannon et al., 2003 | RRID:SCR_003032 | |
| Software, algorithm | GeneMANIA | Warde-Farley et al., 2010 | RRID:SCR_005709 | |
| Software, algorithm | Set Comparison Appyter | Clarke et al., 2021 | RRID:SCR_021245 | |
| Software, algorithm | DGRP2 | Mackay et al., 2012 | http://dgrp2.gnets.ncsu.edu/ |





