Dimer-monomer transition defines a hyper-thermostable peptidoglycan hydrolase mined from bacterial proteome by lysin-derived antimicrobial peptide-primed screening
Figures
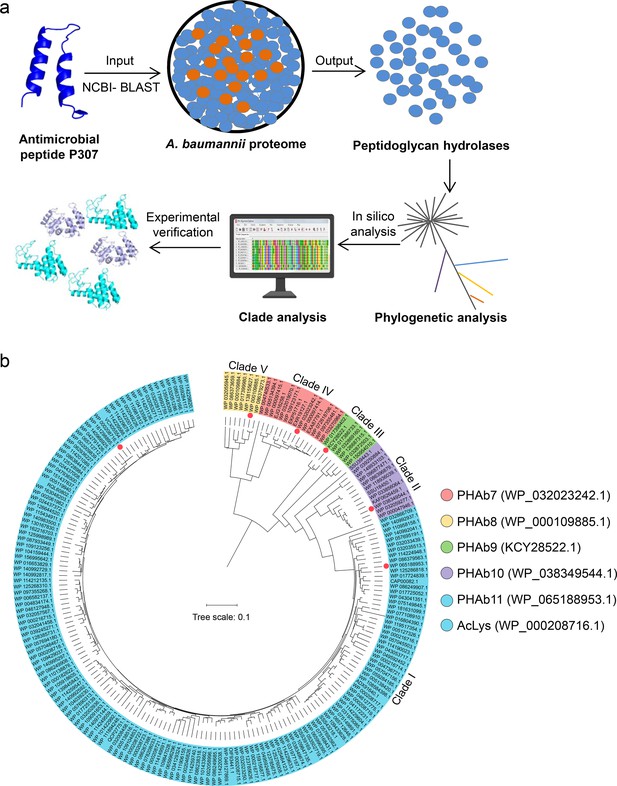
Screening of putative peptidoglycan hydrolases from the A. baumannii proteome.
(a) Workflow for the development of an integrated system for peptidoglycan hydrolase screening. (b) Phylogenetic analysis of putative peptidoglycan hydrolases from the A. baumannii proteome database. Multiple sequence alignments are performed using MEGAX software with ClustalW algorithm, and the neighbor-joining method is used to construct a phylogenetic tree. Classified clades are labeled in different colors and representative peptidoglycan hydrolases from each clade are pointed by colored circles.
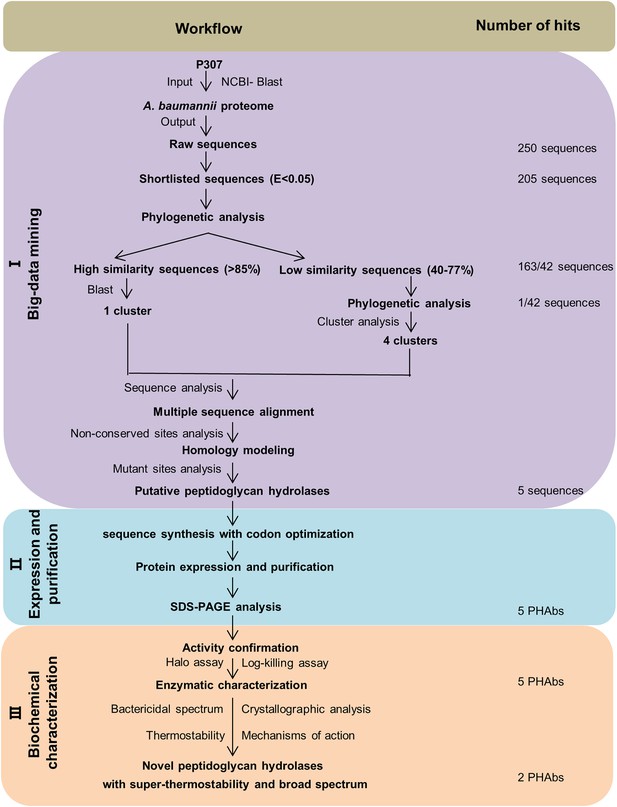
Workflow of the search integrated system for peptidoglycan hydrolases.
The three different stages of the workflow are highlighted in different colors: (I) Screening of peptidoglycan hydrolases using the cationic peptides P307 as a template (yellow). (II) Protein expression and purification (blue). (III) Biochemical characterization (green).
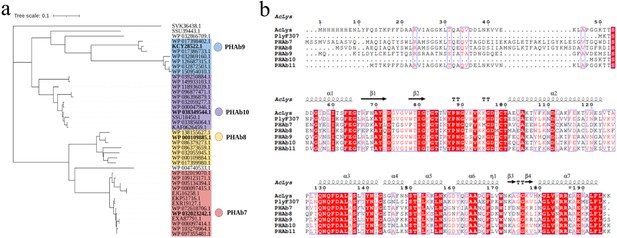
Selective five putative peptidoglycan hydrolases.
(a) Phylogenetic tree of A. baumannii peptidoglycan hydrolases homolog to P307. (b) Multiple sequence alignment of PHAb7, PHAb8, PHAb9, PHAb10, PHAb11, PlyF307, and AcLys using MEGAX software and ClustalW algorithm. Secondary structures are annotated by Espript 3.0 website.
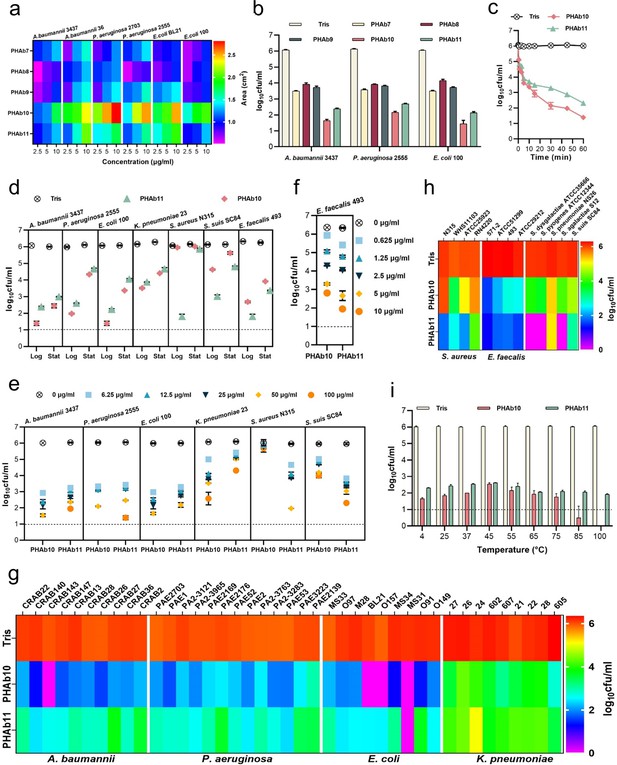
PHAb10 and PHAb11 are highly thermostable peptidoglycan hydrolases with a broad spectrum of action.
(a) Heat map showing the halo size formed by different concentrations of peptidoglycan hydrolases on different bacterial lawns. (b) Bactericidal activity of five peptidoglycan hydrolases (50 μg/ml) in 20 mM Tris-HCl (pH 7.4) at 37°C for 1 hr against different exponential bacteria. (c) Time-dependent bactericidal activity of 50 μg/ml PHAb10 or PHAb11 against exponential A. baumannii in 20 mM Tris-HCl (pH 7.4). (d) Bactericidal activity of 50 μg/ml PHAb10 or PHAb11 in 20 mM Tris-HCl (pH 7.4) at 37°C for 1 hr against different bacteria in exponential and stationary phases. (e–f) Dose-dependent bactericidal activity of PHAb10 and PHAb11 against multiple exponential bacteria in 20 mM Tris-HCl (pH 7.4) at 37°C for 1 hr. (g–h) Susceptibility of various Gram-negative (g) and Gram-positive (h) bacterial strains to PHAb10 and PHAb11. Exponential cultures of each bacterium are treated with 50 μg/ml PHAb10 or PHAb11 for 1 hr at 37°C and residual viable bacterial cells are counted by plating serial dilutions onto agar plates. For Enterococcus faecalis, 10 μg/ml of each peptidoglycan hydrolase is used. (i) Thermal stability of PHAb10 and PHAb11. Each enzyme is stored at different temperatures for 1 hr, cooled to room temperature, and then incubated with exponential A. baumannii 3437 in 20 mM Tris-HCl (pH 7.4) at a final concentration of 50 μg/ml at 37°C for 1 hr. Viable bacteria are counted after each treatment by plating serial dilutions on Lysogeny Broth (LB) agar. All assays were performed with at least three biological replicates (n=3-9). Dash lines represent the limit of detection and data below the limit of detection is not shown.
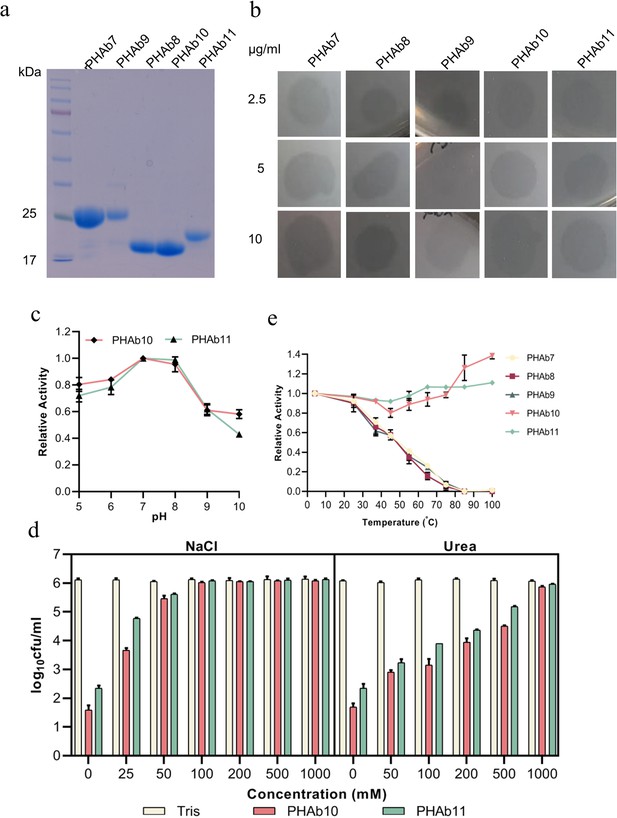
Testing of the bactericidal activity of the screened putative peptidoglycan hydrolases.
(a) Purified proteins were analyzed on a 12% SDS-PAGE gel. M: standard protein marker. (b) Halo of the five PHAbs on a lawn of A. baumannii 3437. The concentrations of each peptidoglycan hydrolase are 2.5 μg/ml, 5 μg/ml, and 10 μg/ml. (c) pH stability of PHAb10 and PHAb11. Log-phase A. baumannii 3437 cells were incubated with 50 μg/ml peptidoglycan hydrolases at 37°C for 1 hr at the indicated pH conditions. pH buffer: 20 mM boric acid and 20 mM phosphoric acid were mixed in equal volumes and adjusted to different pH values with HCl. (d) Log-phase A. baumannii 3437 cells were incubated with 50 μg/ml peptidoglycan hydrolase in 20 mM Tris-HCl (pH 7.4) in the presence of different concentrations of NaCl (left) and urea (right) for 1 hr at 37°C. (e) Relative activity of five PHAbs against A. baumannii 3437 after 1 hr storage at different temperatures. In all cases, viable bacteria were counted by serial dilution and plating assay. Experiments were carried out in triplicate (n=3-9); error bars indicate standard deviation.
-
Figure 2—figure supplement 1—source data 1
PDF file containing original SDS-PAGE gel for Figure 2—figure supplement 1, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/98266/elife-98266-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Original file for SDS-PAGE analysis displayed in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/98266/elife-98266-fig2-figsupp1-data2-v1.zip
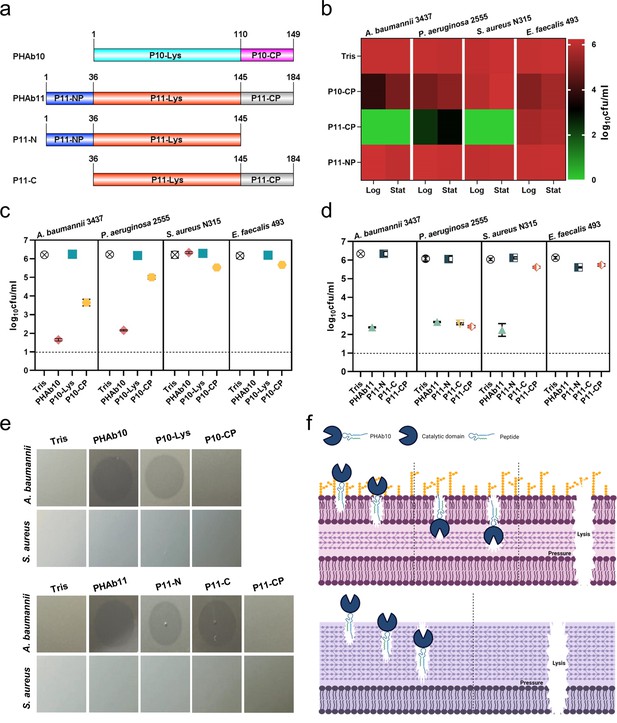
Mechanism of action of PHAb10 and PHAb11 on Gram-negative and Gram-positive bacteria.
(a) Schematic representation of PHAb10 and PHAb11 domains. (b) Bactericidal activity of peptides derived from PHAb10 and PHAb11. Bacterial cells are treated with 50 μg/ml P10-CP, P11-NP, or P11-CP in 20 mM Tris-HCl at 37°C for 1 hr. (c–d) Bactericidal activity of different truncations of PHAb10 and PHAb11. Exponential A. baumannii 3437 cells are incubated with 50 μg/ml of each truncated fragment in 20 mM Tris-HCl for 1 hr at 37°C. Dash lines represent the limit of detection and data below the limit of detection is not shown. (e) Peptidoglycan hydrolytic activity of different truncations of PHAb10 and PHAb11. 0.1 μg of each truncated fragment is dropped onto autoclaved lawns of A. baumannii 3437 and S. aureus N315 and incubated at 37°C for 4 hr. The groups treated with an equal volume of 20 mM Tris-HCl served as controls. (f) Schematic diagram of the bactericidal mechanisms of PHAb10 against Gram-negative bacteria (top) and Gram-positive bacteria (bottom). All assays (b-e) were performed with at least three biological replicates (n=3-9).
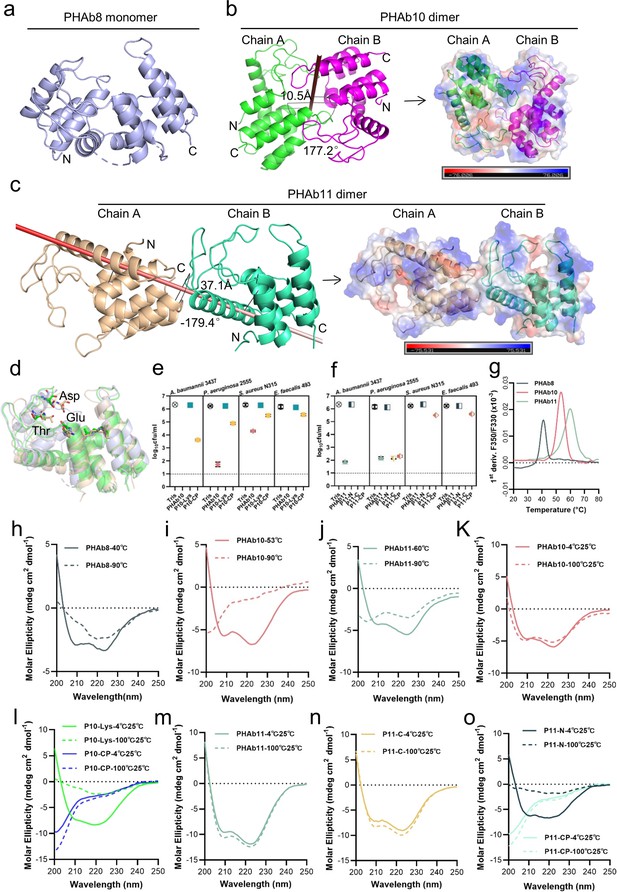
PHAb10 and PHAb11 are thermostable dimers.
(a) Overall structure of PHAb8 monomer. (b) Dimeric structure of PHAb10. Chain A is colored in green, Chain B is colored in magenta, and the gray stick denotes the rotation axis. The electrostatic surface of PHAb10 with two potential substrate-binding cavities are shown (negative = red; positive = blue). (c) Dimeric structure of PHAb11. Chain A is colored in wheat, Chain B is colored in green-cyan, and the red stick denotes the rotation axis. The electrostatic surface of PHAb11 with two potential substrate binding cavities is also shown (negative = red; positive = blue). (d) Superimposition of monomeric PHAb8, PHAb10, and PHAb11. PHAb8 is shown in light blue, PHAb10 in green, and PHAb11 in wheat. Residues involved in the catalytic triad are shown in sticks. (e–f) Residual bactericidal activity of PHAb10, PHAb11, and their truncation variants. Each truncation variant is treated at 100°C for 1 hr, stored at 25°C for 1 hr, and then tested for bactericidal activity by a log-killing assay. All assays were performed with at least three biological replicates (n=3-9). Dash lines represent the limit of detection and data below the limit of detection is not shown. (g) Thermal unfolding curves of PHAb8, PHAb10, and PHAb11 from 20°C to 100°C as determined by nano-differential scanning fluorimetry (nanoDSF). Values on the y-axis represent the first derivative of the fluorescence ratio at 350 nm and 330 nm. Peaks represent the transition temperature of each protein. (h–j) Circular dichroism (CD) spectra of PHAb8 (h), PHAb10 (i), and PHAb11 (j) at 90°C and temperatures close to their transition temperatures. (k–o) CD spectra of PHAb10, PHAb11, and their truncation variants before and after heat treatment. Each domain fragment is treated at 100°C for 1 hr, stored at 25°C for additional hour prior to CD detection.
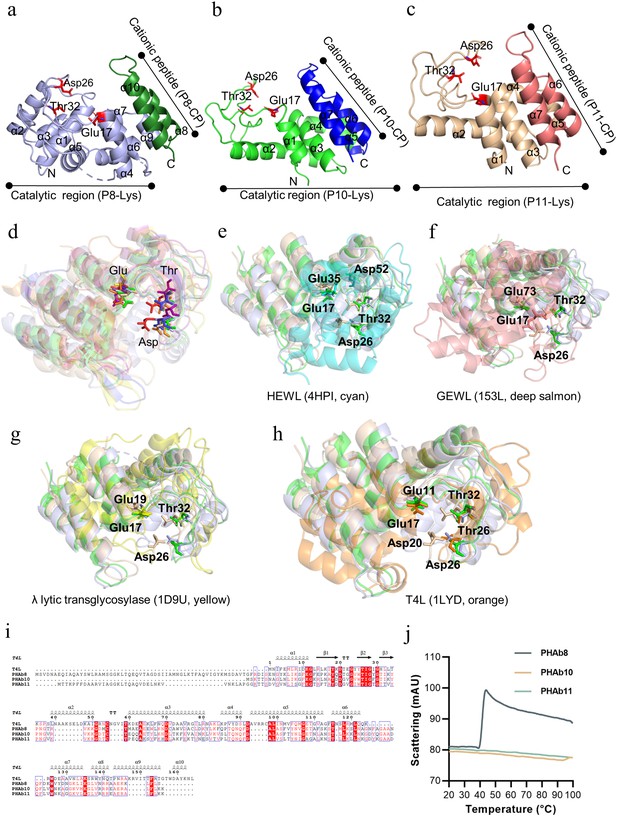
Structural analysis of PHAb8, PHAb10, and PHAb11.
(a–c) Structures of monomeric PHAb8 (a), PHAb10 (b), and PHAb11 (c). (d) Structural superposition of PHAb8, PHAb10, PHAb11, and five classical T4L-like lysozymes that share high similarity from Dali search. PHAb8 is shown in light blue, PHAb10 in green, PHAb11 in wheat, AcLys in red, LysF1 in blue, SpmX-Mur-Ae in yellow, P22 in purple, and R21 in orange. (e–h) Structural alignment of PHAb8, PHAb10, PHAb11 with various lysozymes including hen egg white lysozyme (HEWL) (e, cyan), goose egg white lysozyme (GEWL) (f, deep salmon), phage lambda lytic transglycosylase (g, yellow), and T4 lysozyme (h, orange). Residues in the conserved catalytic triad are shown as sticks. (j) Nano-differential scanning fluorimetry (nanoDSF) scattering plots of PHAb8, PHAb10, and PHAb11 at different temperatures.
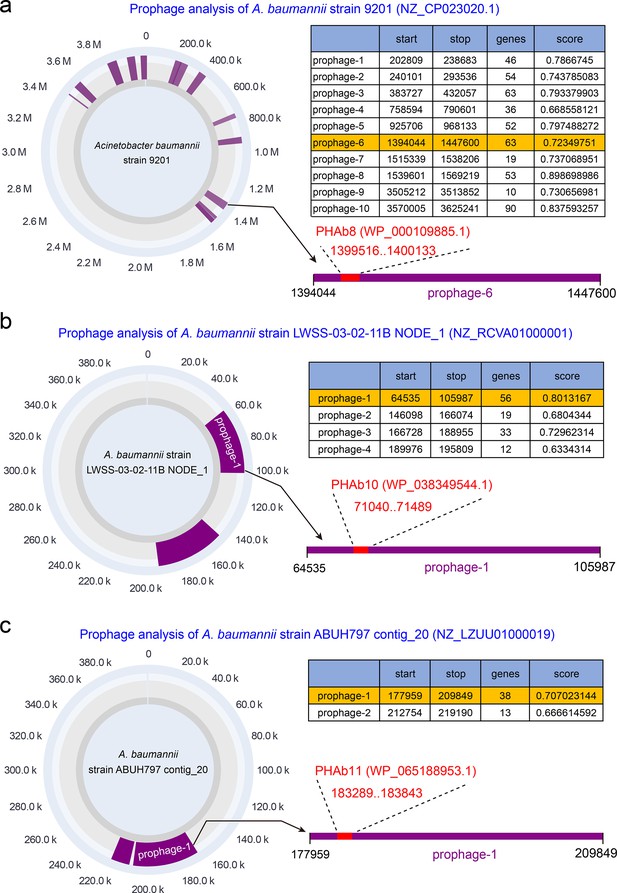
Prophage analysis of PHAbs.
PHAb8, PHAb10, and PHAb11 encoding sequences were retrieved from A. baumannii strain 9201, A. baumannii strain LWSS-03-02-11B, and A. baumannii strain ABUH797 were analyzed by PhageBoost tool. The results show that all three genes are indeed located in the prophage region.
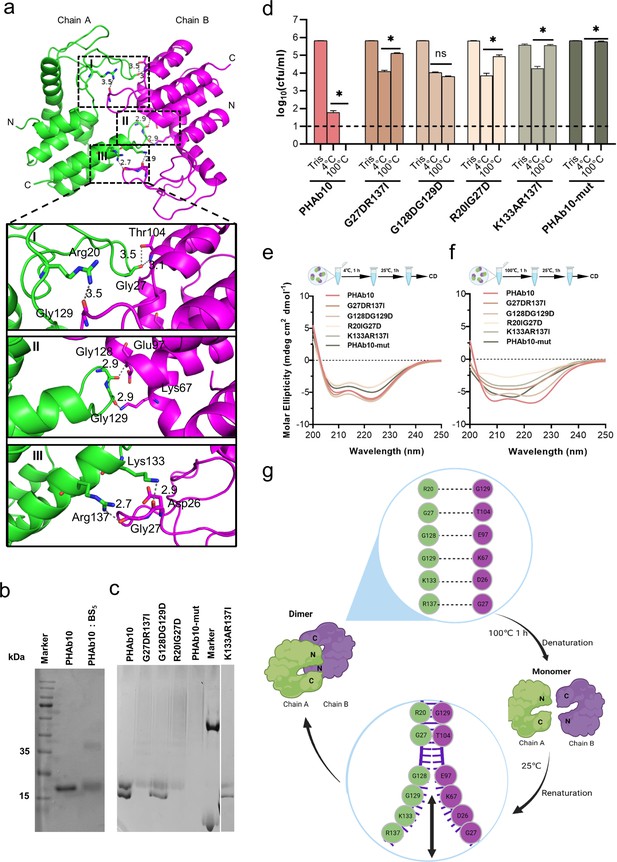
Folding-refolding thermodynamics of PHAb10.
(a) Intermolecular interactions of PHAb10 dimer. The residues involved are shown as sticks. (b) SDS-PAGE analysis of PHAb10 with or without chemical cross-linker. (c) Native-PAGE analysis of PHAb10 and its variants. Marker: the upper band represents BSA (66.4 kDa), and the lower band represents lysozyme (14 kDa). (d) Bactericidal activity of PHAb10 and its variants before and after heat treatment. Each protein is incubated for 1 hr at 4°C or 100°C, stored at 25°C for an additional 1 hr, and then examined for bactericidal activity in 20 mM Tris-HCl (pH 7.4) at a concentration of 50 μg/ml against exponential A. baumannii 3437 cells for 1 hr. All assays were performed with at least three biological replicates (n=6-9). Dash lines represent the limit of detection and data below the limit of detection is not shown. Data are analyzed by two-tailed Student’s t-tests. ns: statistically not significant; *: p<0.05. (e–f) Circular dichroism spectra of PHAb10 and its mutants before and after treatment at 100°C for 1 hr. (g) Zipper model showing the thermodynamics of PHAb10 dimer.
-
Figure 5—source data 1
PDF file containing original SDS/Native-PAGE gels for Figure 5b and c, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/98266/elife-98266-fig5-data1-v1.zip
-
Figure 5—source data 2
Original file for SDS/Native-PAGE analysis displayed in Figure 5b and c.
- https://cdn.elifesciences.org/articles/98266/elife-98266-fig5-data2-v1.zip
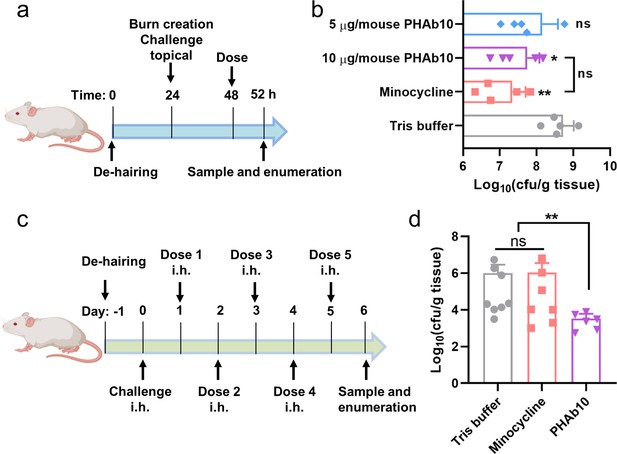
PHAb10 shows high efficacy in mouse infection models.
(a) Experimental schema for burn wound infection model. Burn wounds produced by exposing naked skin to 65°C water for 12 s was infected with 10 µl 5×108 cfu/ml of A. baumannii 3437. At 24 hr post-colonization, mice are treated with 5 or 10 µg/mouse PHAb10 (10 µl; n=5), 4 µg/mouse minocycline (10 µl; n=5), or an equal volume of Tris buffer (10 µl; n=5). Four hours post-treatment, viable bacteria on the burn wound skin is collected and counted. (b) Viable A. baumannii collected from burn wound skin after each treatment. The number of viable cells in each group is normalized and compared with the number of viable cells treated with Tris buffer by one-way analysis of variance (ANOVA). ns: statistically not significant; *: p<0.05; **: p<0.01. (c) Experimental schema for abscess model. Mice are infected hypodermically with 25 μl 5×108 cfu/ml of A. baumannii 3437 on the right side of the back dorsum. Twenty-four hours after infection, mice are injected hypodermically with 10 µg/mouse PHAb10 (10 µl; n=6), 4 µg/mouse minocycline (10 µl; n=7), or an equal volume of Tris buffer (10 µl; n=8). Each group is injected subcutaneously once a day for 5 consecutive days. Viable bacteria were counted at the infection site 24 hr after the last dose. (d) Viable A. baumannii collected from the skin after each treatment. The number of viable cells from each group was normalized and compared with those of the Tris buffer-treated group by ANOVA. ns: statistically not significant; **: p<0.01.
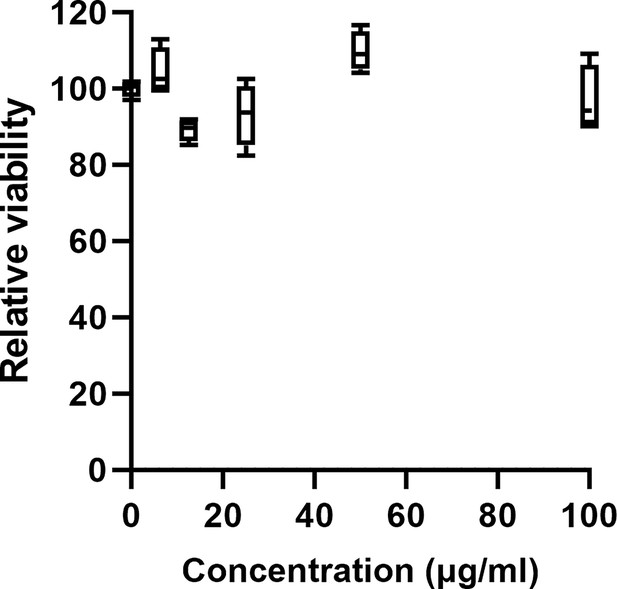
Relative viability of HepG2 cells exposed to different concentrations of PHAb10.
Cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum, 1% penicillin, and 1% streptomycin for 24 hr. After cells were exposed to a series of concentrations of PHAb10 (0, 6.25, 12.5, 25, 50, and 100 μg/ml) for an additional 24 hr, the relative viability of cells after each treatment was determined by Cell Counting Kit-8 (CCK-8) assay. All assays were performed with at least three biological replicates (n=6-9).
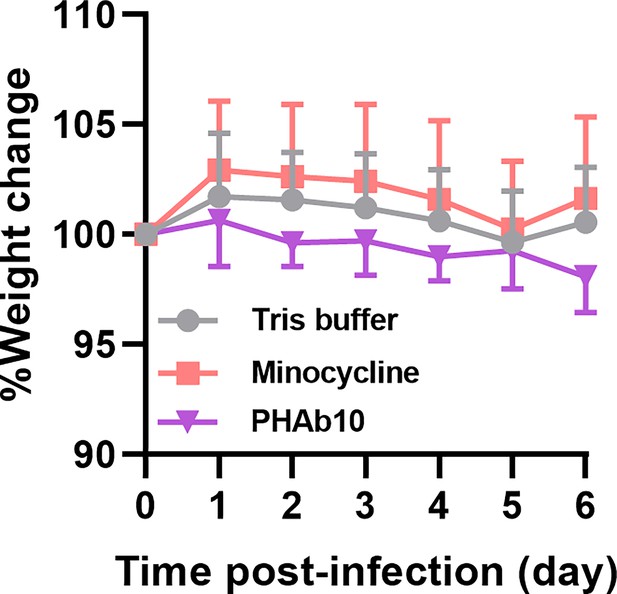
Changes in mouse body weight in a mouse abscess model.
Mice were subcutaneously injected with 25 μl of 5×108 cfu/ml of A. baumannii 3437 on the right side of the back dorsum. Twenty-four hours after infection, mice were injected subcutaneously with 10 µg/mouse PHAb10 (10 µl; n=6), 4 µg/mouse minocycline (10 µl; n=7), or an equal volume of Tris buffer (10 µl; n=8). Each group was subcutaneously injected once a day for 5 consecutive days. The body weight of mice in each group was monitored daily.
Additional files
-
Supplementary file 1
Construction and characterization of PHAbs and their variants.
(a) Physicochemical properties of putative antimicrobial peptides in PHAb10 and PHAb11. (b) Data collection and refinement statistics of PHAb8, PHAb10, and PHAb11 structures. (c) T4L-like lysozymes with characterized structures. (d) Intermolecular interactions of PHAb10 dimer. (e) Design of PHAb10 variants. (f) Bacterial strains used in this work. (g) Primers used in this study.
- https://cdn.elifesciences.org/articles/98266/elife-98266-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98266/elife-98266-mdarchecklist1-v1.docx





