Altering the redox status of Chlamydia trachomatis directly impacts its developmental cycle progression
Figures
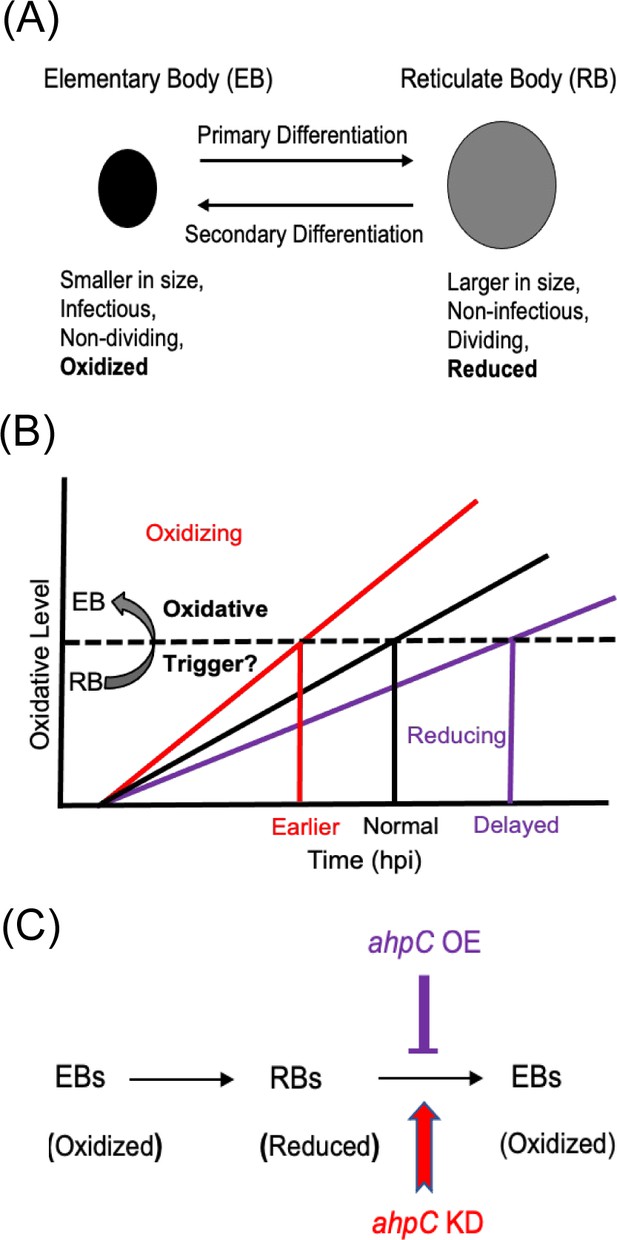
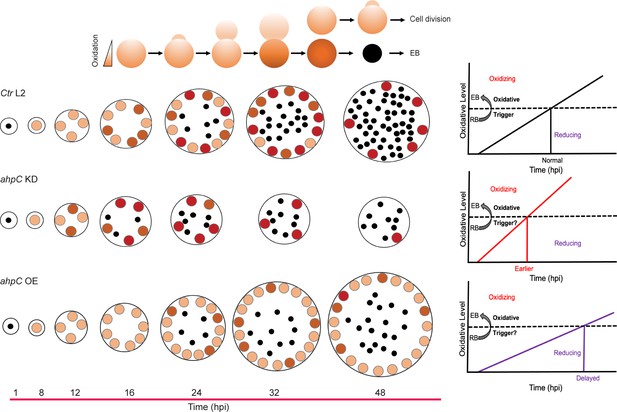
Alterations in the redox status of key proteins regulate and drive chlamydial differentiation.
(A) Key characteristics of chlamydial developmental forms. (B) Hypothetical model for triggering secondary differentiation through oxidative stress (black angled line). Increasing oxidation of critical protein(s) may lead to earlier differentiation whereas maintaining a reducing environment may delay differentiation. (C) Schematic representation of the experimental model for triggering secondary differentiation through the altered activity of alkyl hydroperoxide reductase subunit C (AhpC). ahpC knockdown may lead to earlier differentiation, while overexpression of ahpC may delay differentiation.
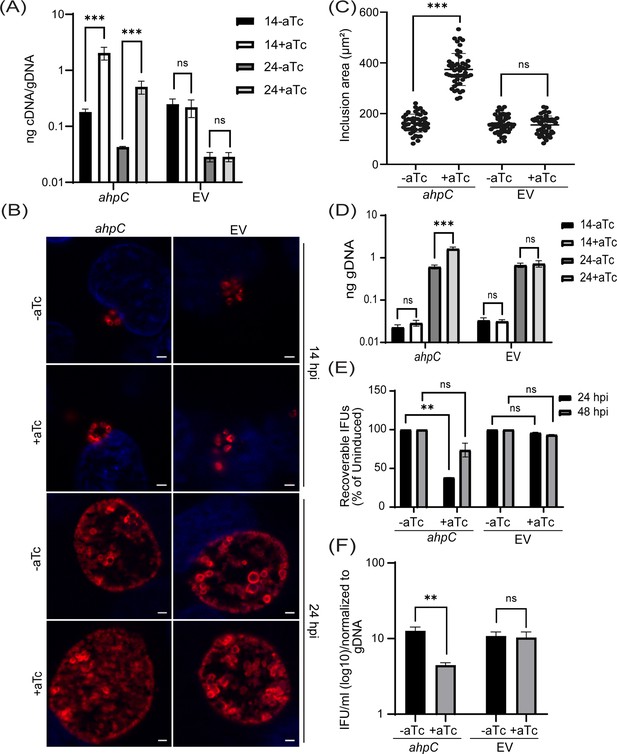
Overexpression of alkyl hydroperoxide reductase subunit C (ahpC) affects chlamydial growth and differentiation.
(A) Transcriptional analysis of ahpC in ahpC overexpression (ahpC) and empty vector (EV) control using RT-qPCR following induction at 10 hpi with 1 nM aTc. RNA and genomic DNA (gDNA) were harvested at 14 and 24 hpi and processed as mentioned in the materials and methods. Data are presented as a ratio of cDNA to gDNA plotted on a log scale. ***p<0.0001 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (B) Immunofluorescence assay (IFA) of ahpC and EV at 14 and 24 hpi. Construct expression was induced or not at 10 hpi with 1 nM aTc, and samples were fixed with methanol at 14 and 24 hpi and then stained for major outer membrane protein (MOMP - red) and DAPI (blue) to label DNA. Scale bars = 2 µm. Images were captured using a Zeiss Axio Imager Z.2 with Apotome2 at 100 x magnification. Representative images of three biological replicates are shown. (C) Impact of ahpC overexpression on inclusion area. Inclusion area of ahpC overexpression and EV strains was measured using ImageJ. Experimental conditions were the same as mentioned in section (B). The area of 50 inclusions was measured per condition for each sample. ***p<0.001 vs uninduced sample by using ordinary one-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data were collected from three biological replicates. (D) Quantification of genomic DNA (gDNA) determined by qPCR in ahpC overexpression and empty vector control. Construct expression was induced or not at 10 hpi with 1 nM aTc, gDNA was harvested at 14 and 24 hpi, and ng gDNA was plotted on a log scale. ***p<0.0001 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (E) IFU assay of ahpC overexpression and empty vector control. Expression of the construct was induced or not at 10 hpi, and samples were harvested at 24 or 48 hpi for reinfection and enumeration. IFUs were calculated as the percentage of uninduced samples. **p<0.001 vs uninduced sample by using multiple paired t-test. Data represent three biological replicates. (F) Ratio of log10 IFUs and log10 gDNA. IFU/ml from (E) was normalized with gDNA from (D). **p<0.001 vs uninduced sample by using multiple unpaired t-test. Data represent three biological replicates.
-
Figure 2—source data 1
RT-qPCR (cDNA and gDNA), quantification of inclusion size, gDNA, and IFU data of ahpC OE and EV strains.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig2-data1-v1.xlsx
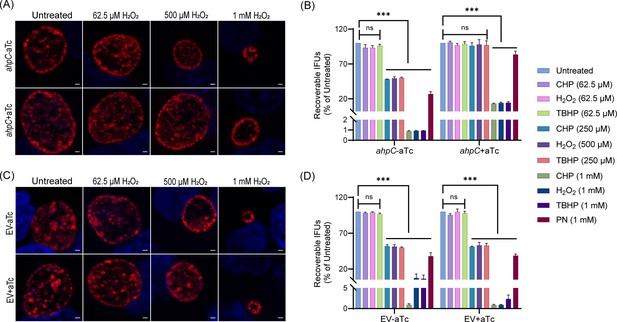
Higher expression of alkyl hydroperoxide reductase subunit C (ahpC) provides resistance to peroxides in Chlamydia.
Immunofluorescence analysis (IFA) of ahpC (A) or empty vector (EV) (C) exposed to oxidizing agents. Construct expression was induced or not at 10 hpi with 1 nM aTc, and samples were treated with three different concentrations of hydrogen peroxide (H2O2) at 16 hpi for 30 min, then fixed with methanol at 24 hpi, stained and imaged as described in the legend of Figure 2B. Representative images from three biological replicates are shown. Scale bars = 2 µm. IFU analysis of ahpC (B) or EV (D) following treatment with oxidizing agents, CHP-Cumene hydroperoxide, H2O2-Hydrogen peroxide, TBHP-Tert-butyl hydroperoxide, and PN-Peroxynitrite. Samples were processed as described for (A) and (C), and IFUs were harvested at 24 hpi. IFUs of treated samples were compared with respective untreated controls. ***p<0.0001 vs untreated sample by using two-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates.
-
Figure 3—source data 1
IFU data of ahpC OE and EV against CHP, hydrogen peroxide, TBHP, and PN.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig3-data1-v1.xlsx
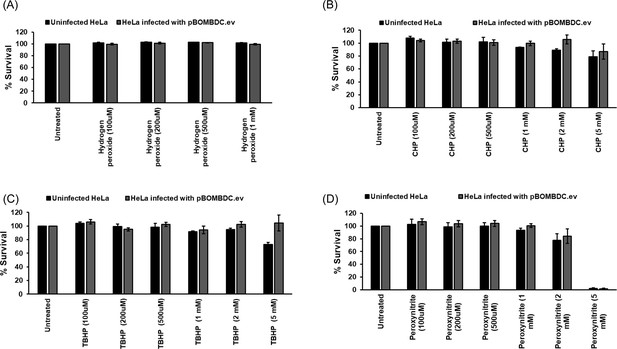
Viability assay of uninfected or infected HeLa cells treated with oxidizing agents hydrogen peroxide (H2O2) (A), cumene hydroperoxide (CHP) (B), tert-butyl hydroperoxide (TBHP) (C), and peroxynitrite (PN) (D).
HeLa cells were infected or not with empty vector control, pBOMBDC.ev (empty vector, EV), and treated or not with different concentrations of oxidizing agents at 16 hpi for 30 min. At 24 hpi, an end point viability assay was performed using PrestoBlue as mentioned in materials and methods. The treated values were expressed as a percentage of the untreated values, which were considered as 100%. Data represent three biological replicates.
-
Figure 3—figure supplement 1—source data 1
Viability data of uninfected or infected HeLa cells in presence of hydrogen peroxide, CHP, TBHP, and PN.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig3-figsupp1-data1-v1.xlsx
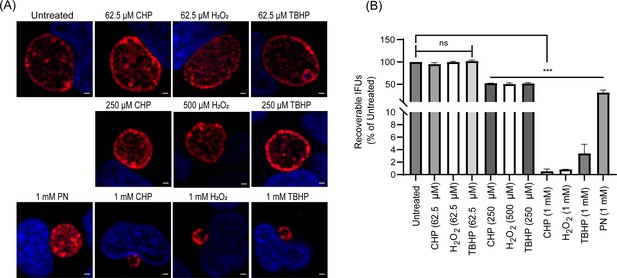
Response of C.trachomatis L2 against oxidizing agents.
(A) Immunofluorescence analysis (IFA) of wild-type Ctr L2 exposed to oxidizing agents. Oxidizing agents’ treatment, staining, and imaging were performed as mentioned in the materials and methods. Representative images from three biological replicates are shown. Scale bars = 2 µm. (B) IFU analysis of Ctr L2 post-exposure with oxidizing agents. Conditions were the same as in section (A), and IFUs were harvested at 24 hpi. IFUs were calculated as a percentage of untreated samples. ***p<0.0001 vs untreated sample by using one-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates.
-
Figure 3—figure supplement 2—source data 1
IFU data of Ctr L2 against CHP, hydrogen peroxide, TBHP, and PN.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig3-figsupp2-data1-v1.xlsx
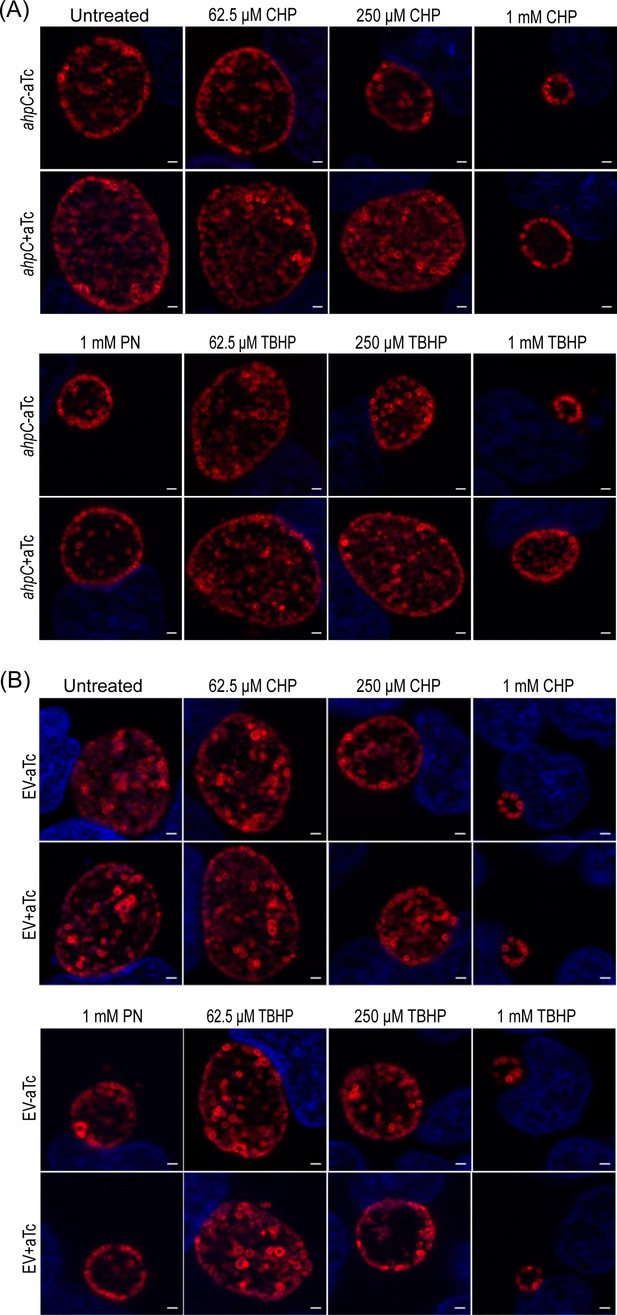
Overexpression of alkyl hydroperoxide reductase subunit C (ahpC) provides resistance to peroxides in Chlamydia.
Immunofluorescence analysis (IFA) of ahpC (A) or empty vector (EV) (B) exposed to oxidizing agents (cumene hydroperoxide CHP, tert-butyl hydroperoxide TBHP, and peroxynitrite (PN)). Experiments were performed as mentioned in the legend of Figure 3A. Representative images from three biological replicates are shown. Scale bars = 2 µm.
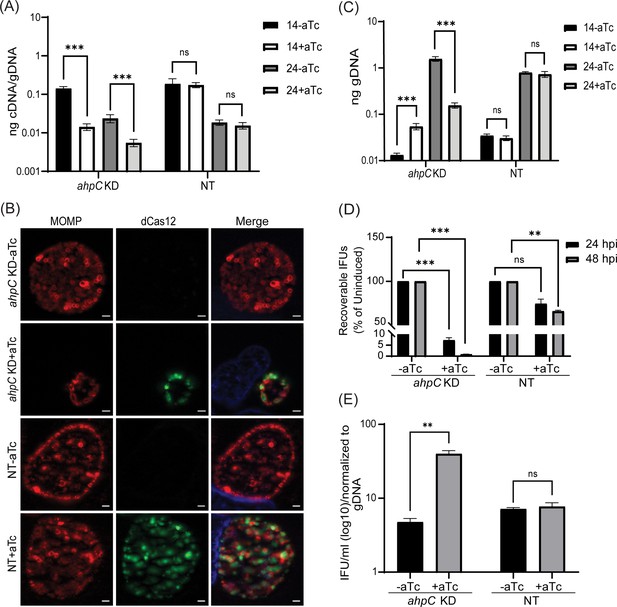
Reduced levels of alkyl hydroperoxide reductase subunit C (AhpC) negatively impact chlamydial growth.
(A) Transcriptional analysis of ahpC in knockdown (ahpC KD) and non-target (NT) control using RT-qPCR following induction at 10 hpi with 1 nM aTc. RNA and gDNA were harvested at 14 and 24 hpi. Quantified cDNA was normalized to gDNA, and values were plotted on a log scale. ***p<0.0001 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (B) Immunofluorescence analysis (IFA) was performed to assess inclusion size and morphology using the same induction conditions as in section (A). At 24 hpi, cells were fixed with methanol and stained using primary antibodies to major outer membrane protein (MOMP), Cpf1 (dCas12), and DAPI. All images were acquired on Zeiss Axio Imager Z.2 with Apotome2 at 100 x magnification. Scale bars = 2 µm. Representative images of three biological replicates are shown. (C) Quantification of genomic DNA (gDNA) determined by qPCR in ahpC KD and NT strains. dCas12 expression was induced or not at 10 hpi, and gDNA was harvested at 14 and 24 hpi and plotted on a log scale. ***p<0.0001 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (D) IFU titers following induction at 10 hpi with 1 nM aTc. IFUs were counted from 24 and 48 hpi samples and calculated as a percentage of uninduced samples. ***p<0.0001, **p<0.001 vs uninduced sample by using multiple paired t-test. Data represent three biological replicates. (E) Ratio of log10 IFUs by log10 gDNA. IFU/ml from (D) was normalized with gDNA from (C). **p<0.001 vs uninduced sample by using multiple unpaired t-test. Data represent three biological replicates.
-
Figure 4—source data 1
RT-qPCR (cDNA and gDNA), gDNA, and IFU data of ahpC KD and NT strains.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig4-data1-v1.xlsx
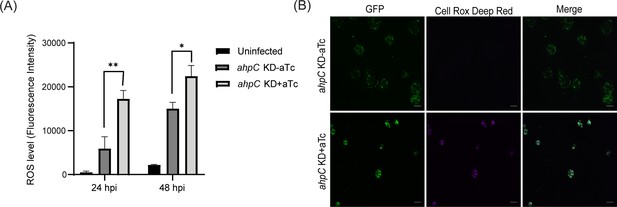
Intracellular reactive oxygen species (ROS) levels were measured to investigate the function of alkyl hydroperoxide reductase subunit C (AhpC) in reducing ROS.
(A) HeLa cells were infected or not with ahpC knockdown, and the construct was induced or not at 10 hpi with 1 nM aTc. At 24 or 48 hpi, samples were washed with DPBS and incubated with CellROX Deep red dye for 30 min in the dark. ROS levels were measured at wavelengths of 640 nm (excitation) and 665 nm (emission). **p<0.001, *p<0.01 vs uninduced sample by using two-way ANOVA, and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (B) Microscopy images were acquired using live cells on Zeiss Axio Imager Z.2 with Apotome2 at 100 x magnification. Scale bars = 10 µm. Representative images of three biological replicates are shown.
-
Figure 5—source data 1
Values of ROS measurement in uninfected HeLa cells and ahpC KD strain in the uninduced and induced conditions.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig5-data1-v1.xlsx
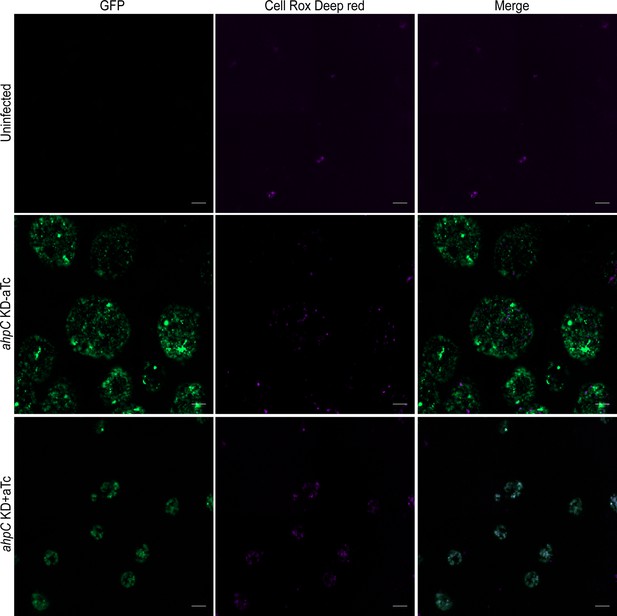
Intracellular reactive oxygen species (ROS) levels in alkyl hydroperoxide reductase subunit C (ahpC) knockdown at 40 hpi.
HeLa cells were infected or not with ahpC knockdown, and the construct was induced or not at 10 hpi with 1 nM aTc. At 40 hpi, samples were washed with DPBS and incubated with CellROX Deep red dye for 30 min in the dark. Microscopic images of live cells on Zeiss Axio Imager Z.2 with Apotome2 at 100 x magnification were captured. Scale Bars = 10 µm. Representative images of three biological replicates are shown.
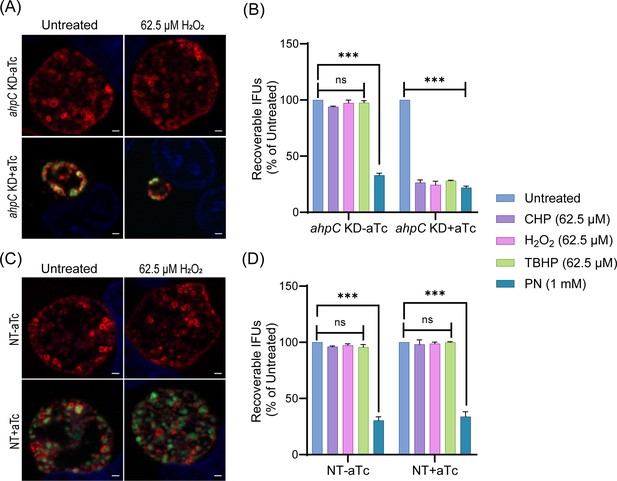
Chlamydia is hypersensitive to oxidizing agents in alkyl hydroperoxide reductase subunit C (ahpC) knockdown condition.
Immunofluorescence analysis (IFA) of ahpC KD (A) or NT (C) treated with 62.5 µM H2O2. dCas12 expression was induced or not at 10 hpi with 1 nM aTc, treated or not with H2O2 at 16 hpi for 30 min, and allowed to grow until 24 hpi. Coverslips were fixed with methanol at 24 hpi and stained major outer membrane protein (MOMP), Cpf1 (dCas12), and DAPI. Scale bars = 2 µm. Images were captured using a Zeiss Axio Imager Z.2 with Apotome2 at 100 x magnification. Representative images from three biological replicates are shown. IFU analysis of ahpC KD (B) or NT (D) following treatment with oxidizing agents, CHP-Cumene hydroperoxide, H2O2-Hydrogen peroxide, TBHP-Tert-butyl hydroperoxide, or PN-Peroxynitrite. dCas12 expression was induced or not, and samples were treated or not as mentioned in the legend of Figure 3B. IFUs of treated samples were calculated as a percentage of respective untreated samples. ***p<0.0001 vs untreated sample by using two-way ANOVA and Tukey’s HSD was applied as post hoc test. Data represent three biological replicates.
-
Figure 6—source data 1
IFU values after CHP, hydrogen peroxide, TBHP, and PN treatment in the ahpC KD and NT strains.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig6-data1-v1.xlsx
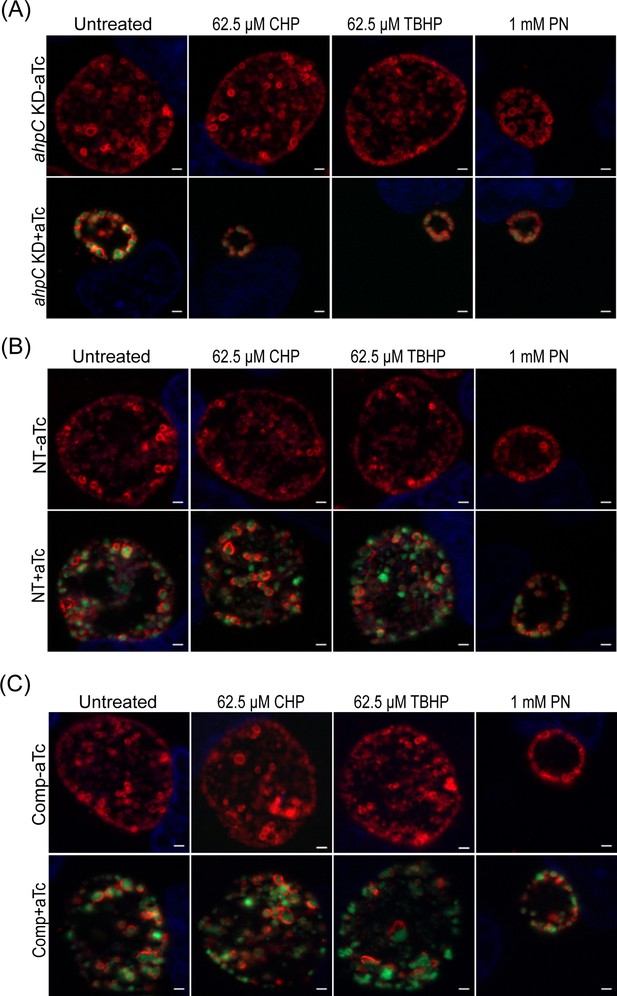
Chlamydia is hypersensitive to oxidizing agents as a result of reduced levels of alkyl hydroperoxide reductase subunit C (ahpC).
Immunofluorescence analysis (IFA) of ahpC KD (A), NT (B), comp (C) treated with cumene hydroperoxide (CHP), tert-butyl hydroperoxide (TBHP), and peroxynitrite (PN). Experimental conditions were the same as mentioned in the legend of Figure 6A. Representative images from three biological replicates are shown. Scale bars = 2 µm.
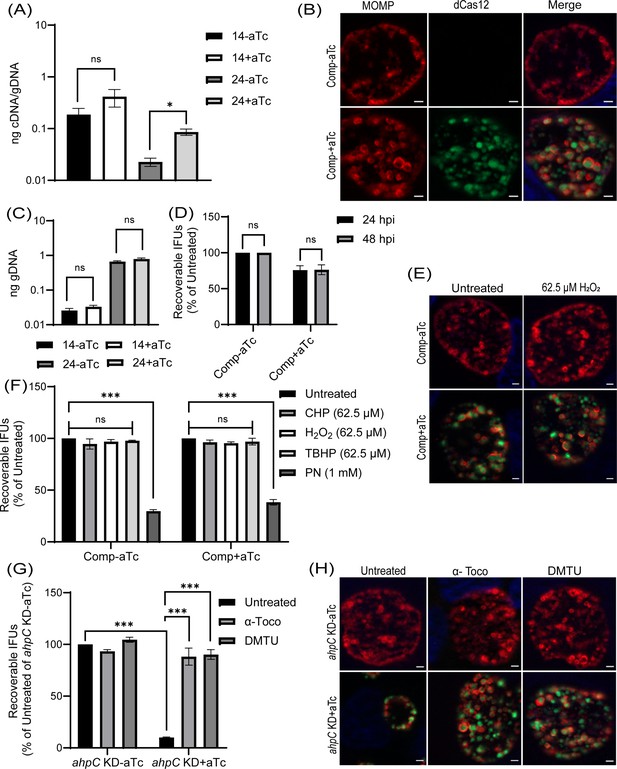
Complementation of the phenotypes observed in the alkyl hydroperoxide reductase subunit C (ahpC) knockdown.
(A) Confirmation of complementation (comp) of ahpC knockdown by RT-qPCR. Samples were processed and quantified as mentioned previously in the legend of Figure 4A. Values were plotted on a log scale. *p<0.01 vs uninduced sample using ordinary one-way ANOVA and Tukey HSD was applied as post hoc test. Data represent three biological replicates. (B) Immunofluorescence analysis (IFA) of comp strain was performed at 24 hpi, and staining and imaging was performed as mentioned in the legend of Figure 4B. Scale bars = 2 µm. Representative images from three biological replicates are shown. (C) Genomic DNA quantitation was performed by qPCR. Construct expression was induced or not at 10 hpi, and gDNA was harvested at 14 and 24 hpi and plotted on a log scale. Statistical analysis was calculated using ordinary one-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (D) IFU analysis of comp strain. Statistical analysis was calculated using multiple paired t-test. Data represent three biological replicates. (E) IFA of comp strain following treatment with 62.5 µM H2O2. Samples were treated, stained, and images acquired as mentioned in the legend of Figure 6A. Scale bars = 2 µm. Representative images from three biological replicates are shown. (F) IFU analysis of comp strain following treatment with oxidizing agents. Experiments were performed as mentioned in the legend of Figure 6B. IFUs were calculated as a percentage of respective untreated samples. ***p<0.0001 vs untreated sample by using two-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (G) ahpC knockdown growth defect rescued by ROS scavengers. IFU analysis of ahpC knockdown treated with or without scavengers, α-Tocopherol (100 µM) and DMTU (10 mM), as mentioned in materials and methods. IFUs were calculated as a percentage of the untreated, uninduced sample. ***p<0.0001 vs untreated, uninduced, or induced sample by using two-way ANOVA and Tukey’s HSD was applied as a post hoc test. Data represent three biological replicates. (H) IFA of ahpC knockdown treated with or without scavengers. Experimental conditions were similar as in section (G). Staining and imaging were performed as mentioned in Figure 6A. Representative images from three biological replicates are shown. Scale bars = 2 µm.
-
Figure 7—source data 1
RT-qPCR (cDNA and gDNA), gDNA, and IFU after hydrogen peroxide stress in the complemented strain and IFU data in presence of scavengers in the ahpC KD strain.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig7-data1-v1.xlsx
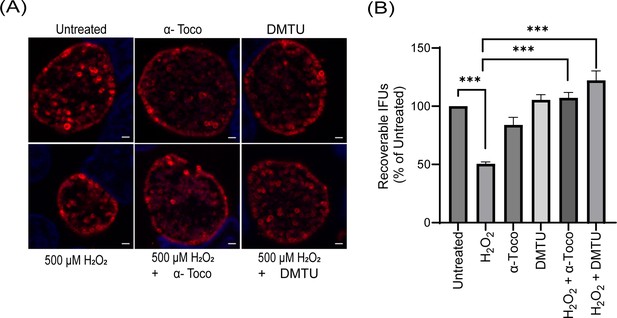
Rescue of oxidative stress phenotype by reactive oxygen species (ROS) scavengers in C. trachomatis L2.
(A) Immunofluorescence analysis (IFA) of wild-type Ctr L2 incubated or not with scavengers, α-Tocopherol (100 µM), and DMTU (10 mM), and treated or not with 500 µM H2O2 as mentioned in materials and methods. Representative images from three biological replicates are shown. Scale bars = 2 µm. (B) IFU analysis of Ctr L2 grown under the same conditions as mentioned in the legend of Figure 7G. IFUs were calculated as a percentage of untreated samples. ***p<0.0001, statistical analysis was performed using ordinary one-way ANOVA, and Tukey’s HSD was applied as post hoc test. H2O2-treated samples and samples incubated with scavengers only were compared to the untreated control. Samples treated with H2O2 and incubated with scavengers were compared with sample treated with H2O2. Data represent three biological replicates. Only significant differences are noted.
-
Figure 7—figure supplement 1—source data 1
IFU data of Ctr L2 in response to scavengers addition after treatment with hydrogen peroxide.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig7-figsupp1-data1-v1.xlsx
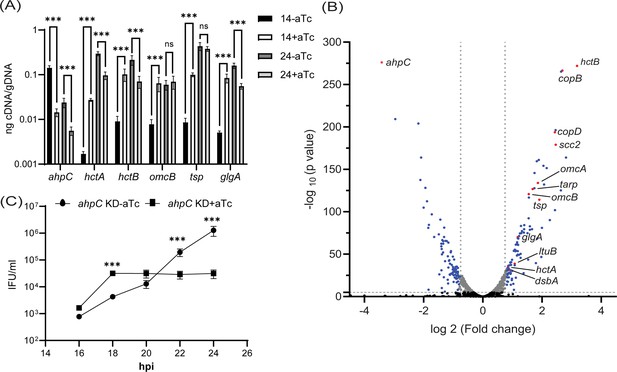
Effects of alkyl hydroperoxide reductase subunit C (ahpC) knockdown on chlamydial developmental cycle progression.
(A) RT-qPCR analysis of late-cycle genes (hctA, hctB, omcB, tsp, and glgA) in ahpC knockdown. Experimental conditions were the same as mentioned in the legend of Figure 4A. Quantified cDNA was normalized to gDNA, and values were plotted on a log scale. ***p<0.0001 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as post hoc test. Data represent three biological replicates. (B) Volcano plot of RNA-sequencing of ahpC knockdown. Experimental conditions were the same as mentioned in the legend of Figure 4A. The volcano plot was prepared using GraphPad Prism software. The vertical dashed lines indicate a fold change of 1.5 as compared to the respective uninduced control. The horizontal dashed line indicates a pvalue of 0.05. The black dots represent genes not significantly different and gray dots represent significantly altered but fold change lesser than 1.5. Blue and red spots represent statistically significant altered genes with more than a 1.5-fold change in transcription levels between uninduced and induced samples. Red dots indicate ahpC and canonical late genes also shown in Table 1. (C) One-step growth curve of ahpC knockdown. Samples were induced or not with 1 nM aTc at 10 hpi and harvested at 16, 18, 20, 22, and 24 hpi. IFUs recovered are displayed as log10 values. ***p<0.0001 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as post hoc test. Data represent three biological replicates.
-
Figure 8—source data 1
RT-qPCR (cDNA, gDNA), volcano plot of RNA-seq, and one step growth curve data of ahpC KD.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig8-data1-v1.xlsx
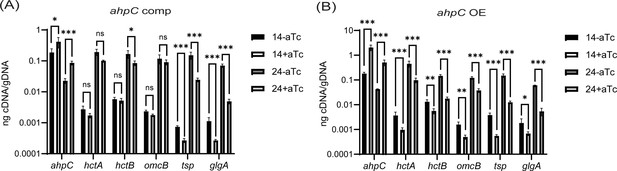
Effects of alkyl hydroperoxide reductase subunit C (ahpC) knockdown/overexpression on chlamydial developmental cycle progression.
RT-qPCR analysis of late-cycle genes (hctA, hctB, omcB, tsp, and glgA) in (A) complementation, and (B) ahpC overexpression strain. Experimental conditions were the same as mentioned in the legend of Figures 7A and 2A, respectively. Quantified cDNA was normalized to gDNA, and values were plotted on a log scale. ***p<0.0001, **p<0.001, *p<0.01 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as post hoc test. Data represent three biological replicates.
-
Figure 8—figure supplement 1—source data 1
RT-qPCR (cDNA and gDNA) data of complemented and ahpC OE strains.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig8-figsupp1-data1-v1.xlsx
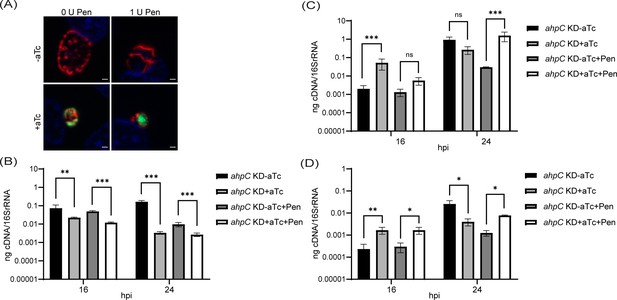
Effect of penicillin treatment during alkyl hydroperoxide reductase subunit C (ahpC) knockdown.
(A) Immunofluorescence analysis (IFA) was performed to assess inclusion size and morphology of ahpC KD-spec following induction (1 nM aTc) and penicillin treatment (1 U/mL) at 10 hpi. At 24 hpi, cells were fixed with methanol and stained using primary antibodies to major outer membrane protein (MOMP), Cpf1 (dCas12), and DAPI. All images were acquired on Zeiss Axio Imager Z.2 with Apotome2 at 100 x magnification. Scale bars = 2 µm. Representative images of three biological replicates are shown. Transcriptional analysis of (B) ahpC, (C) hctA, and (D) hctB in ahpC KD-spec using RT-qPCR using the same conditions as in section (A). RNA samples were harvested at 16 and 24 hpi. Quantified cDNA was normalized to 16 S rRNA, and values were plotted on a log scale. ***p<0.0001, **p<0.001, *p<0.01 vs uninduced sample by using two-way ANOVA and Tukey’s HSD was applied as post hoc test. Data represent three biological replicates.
-
Figure 8—figure supplement 2—source data 1
RT-qPCR (cDNA and gDNA) data in the ahpC KD-spec strain.
- https://cdn.elifesciences.org/articles/98409/elife-98409-fig8-figsupp2-data1-v1.xlsx
Altering the activity of alkyl hydroperoxide reductase subunit C (AhpC) in Ctr L2 impacts its developmental cycle progression.
(Top) In Chlamydia, secondary differentiation is asynchronous and reticulate bodies (RBs) divide through an asymmetric budding mechanism. In such conditions, either the mother or daughter cell may inherit more oxidized proteins (represented by a darker shade), which can then impact whether a given RB will divide again or undergo secondary differentiation. (Bottom) The black dots represent EBs, the orange circles show RBs. The developmental cycle of wild-type C. trachomatis (Ctr L2) is shown. In ahpC KD, highly oxidized conditions lead some RBs to cross the oxidative threshold sooner, allowing activation of late genes and secondary differentiation earlier than other RBs. In ahpC overexpression, a reducing environment results in a delay in achieving the oxidative threshold, thus allowing RBs to continue to divide before committing to secondary differentiation.
Tables
List of canonical late genes significantly increased during ahpC knockdown at 14 hpi.
| CT # | CTL # | Name | Function | Fold change | Ref |
|---|---|---|---|---|---|
| CT046 | CTL0302 | hct2 (hctB) | Histone H1-like protein HC2 | 9.10 | Brickman and Hackstadt, 1993 |
| CT578 | CTL0841 | copB | Needle tip; translocator | 6.44 | Ouellette et al., 2005 |
| CT576 | CTL0839 | scc2 (lcrH_1) | Type III secretion chaperone (Low calcium response protein H) | 5.52 | Ouellette et al., 2005 |
| CT579 | CTL0842 | copD | Needle tip; translocator | 5.41 | Ouellette et al., 2005 |
| CT441 | CTL0700 | tsp | Carboxy-terminal processing protease | 3.76 | Swoboda et al., 2023 |
| CT444 | CTL0703 | omcA | Small cysteine-rich outer membrane protein | 3.63 | Hatch, 1996 |
| CT456 | CTL0716 | tarP | Translocated actin-recruiting phosphoprotein | 3.22 | Clifton et al., 2004 |
| CT443 | CTL0702 | omcB | Large cysteine-rich periplasmic protein | 2.94 | Newhall, 1987 |
| CT798 | CTL0167 | glgA | Glycogen synthase | 2.26 | Belland et al., 2003; Gehre et al., 2016 |
| CT080 | CTL0336 | ltuB | Late transcription unit B protein | 2.11 | Belland et al., 2003 |
| CT177 | CTL0429 | dsbA | Disulfide bond chaperone | 1.91 | Christensen et al., 2019 |
| CT743 | CTL0112 | hctA | Histone H1-like protein HC1 | 1.81 | Barry et al., 1993 |
Additional files
-
Supplementary file 1
Summary of alkyl hydroperoxide reductase subunit C (ahpC) knockdown RNA-seq results.
- https://cdn.elifesciences.org/articles/98409/elife-98409-supp1-v1.xlsx
-
Supplementary file 2
List of plasmids, strains, and primers used in this study.
- https://cdn.elifesciences.org/articles/98409/elife-98409-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98409/elife-98409-mdarchecklist1-v1.docx





