Virus adaptation to heparan sulfate comes with capsid stability tradeoff
Figures
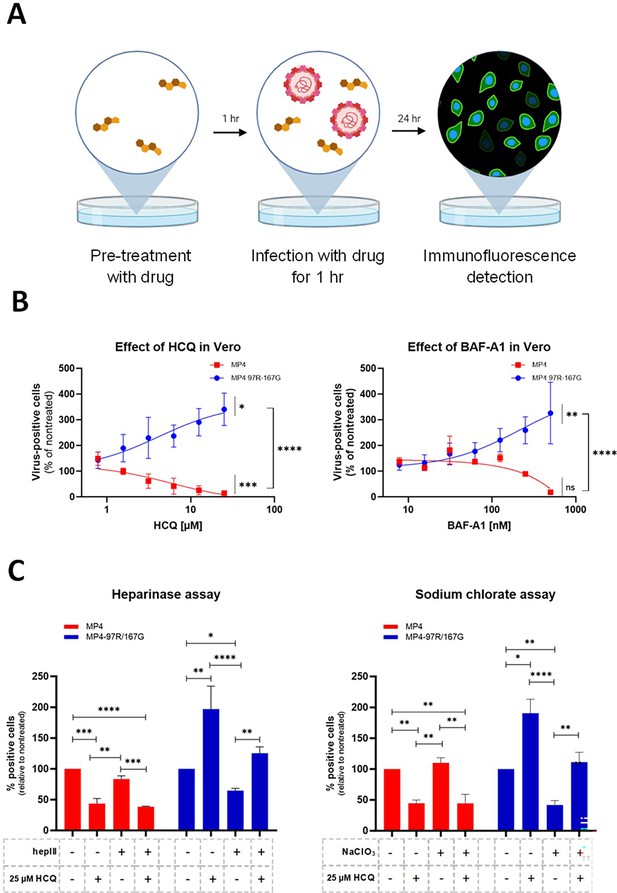
Lysosomotropic drugs inhibit infection by MP4 but not by MP4-97R/167G.
(A) Schematic illustration of the virus inhibitory assay workflow. Cells were pre-treated with lysosomotropic drugs and infected (MOI 0.1) in presence of the drug. After inoculum removal, infected cells were cultured in drug-free media and infected cells were stained by immunofluorescence (IF) with anti-VP2 Ab. Figure 1A was created with BioRender.com. (B) Hydroxychloquine (HCQ) and Bafilomycin A1 (BAF-A1) dose response assay in infected Vero cells. (C) HCQ effect in Vero cells pre-treated or not with heparinase III (hepIII) or sodium chlorate (NaClO3) as in A. Mean and S.E.M of biological triplicates are shown. Results are shown as % of virus-positive cells relative to nontreated control. In B, statistical significance (one-way ANOVA) between treated and untreated virus or between treated MP4 and MP4-97R/167G was calculated based on the area under curve (AUC). In C, statistical significance (two-way ANOVA) was calculated for each virus between each condition. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 1—source data 1
Related to Figure 1B.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Related to Figure 1C.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-data2-v3.xlsx
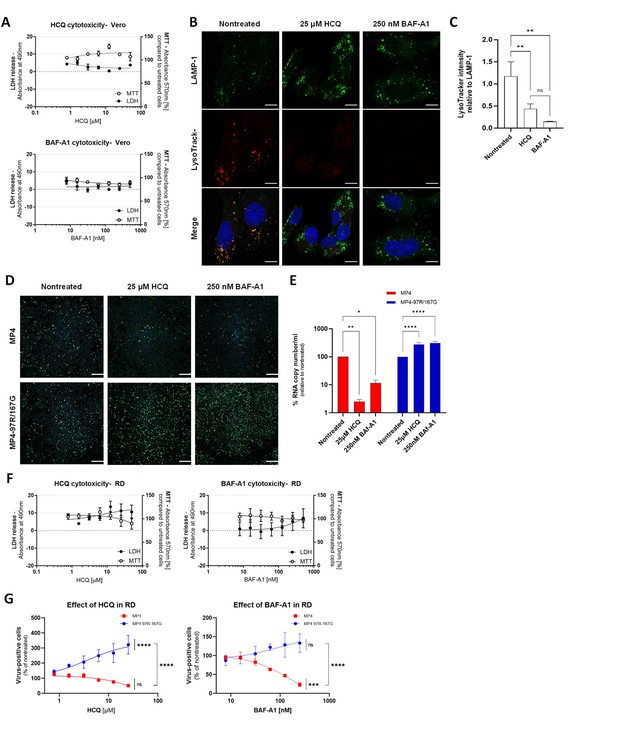
Toxicity and efficacy of lysosomotropic drugs.
(A) The cytotoxic effect of lysosomotropic drugs was evaluated with LDH and MTT assays. Vero cells were treated with a range of different concentrations of hydroxychloroquine (HCQ) or bafilomycin A1 (BAF-A1) for 2 hr. At 24 hr post-treatment, cell supernatants and lysates were collected for LDH assay and MTT assay, respectively, to determine cytotoxicity effect (n=2 biological replicates). (B) Inhibition of endosomal acidification confirmed with LysoTracker staining (red). Lysosomes are stained in green with an anti-LAMP-1 Ab and nuclei in blue (DAPI). Representative immunofluorescence (IF) images are shown (scale bar, 10 µm). (C) Quantified LysoTracker signal intensities calculated from nontreated, HCQ and BAF-A1-treated conditions are shown relative to lysosomal-associated membrane protein 1 (LAMP-1) intensity (n=3 biological replicates). Statistical significance was calculated with one-way ANOVA. (D) Representative IF staining of EV-A71 (anti-VP2 in green) 24 hpi of Vero cells in presence of 25 µM HCQ or 250 nM BAF-A1 (scale bar, 300 µm). (E) Viral RNA load quantification by real-time RT-qPCR of infected Vero cells with and without drug treatment at 24 hpi. (F) Cytotoxic effect of HCQ and BAF-A1 were evaluated using LDH and MTT assays in Rhabdomyosarcoma (RD) cells. (G) Dose response assay with HCQ and BAF-A1 on RD cells were performed exactly like in Vero cells (Figure 1). Infected cells (stained with anti-VP2 Ab) were quantitated at 24 hpi after treatment with increasing drug concentrations. Results are shown as % of virus-positive cells relative to nontreated control. Area under curve (AUC) was calculated and statistical significance (one-way ANOVA) between treated and untreated virus or between treated MP4 and MP4-97R/167 G are shown. Mean and S.E.M of biological duplicates are shown. **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 1—figure supplement 1—source data 1
Related to Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-figsupp1-data1-v3.xlsx
-
Figure 1—figure supplement 1—source data 2
Related to Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-figsupp1-data2-v3.xlsx
-
Figure 1—figure supplement 1—source data 3
Related to Figure 1—figure supplement 1E.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-figsupp1-data3-v3.xlsx
-
Figure 1—figure supplement 1—source data 4
Related to Figure 1—figure supplement 1F.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-figsupp1-data4-v3.xlsx
-
Figure 1—figure supplement 1—source data 5
Related to Figure 1—figure supplement 1G.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-figsupp1-data5-v3.xlsx
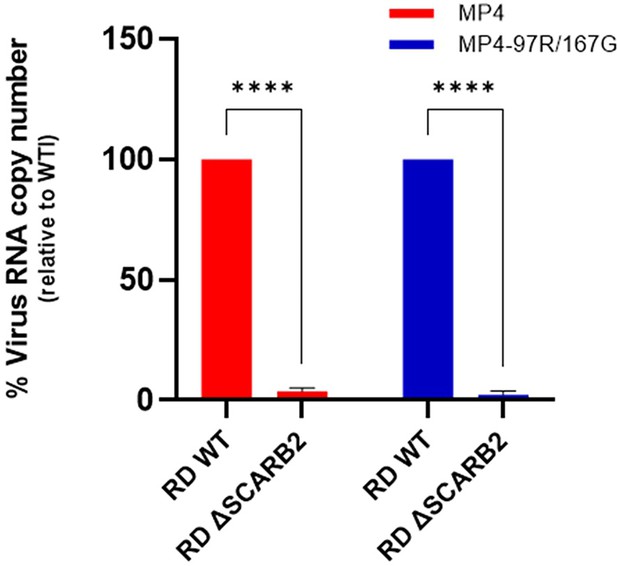
Virus infection in SCARB2-KO cells.
Virus infection was performed on Rhabdomyosarcoma (RD) WT and RD ΔSCARB2 cells. Cells were lysed, and viral RNA copy numbers were quantitated at 24 hpi using RT-qPCR. Results are expressed as % Virus RNA copy number relative to RD WT cells (set to 100%). Mean and S.E.M of biological triplicates are shown.
****p<0.0001.
-
Figure 1—figure supplement 2—source data 1
Related to Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig1-figsupp2-data1-v3.xlsx
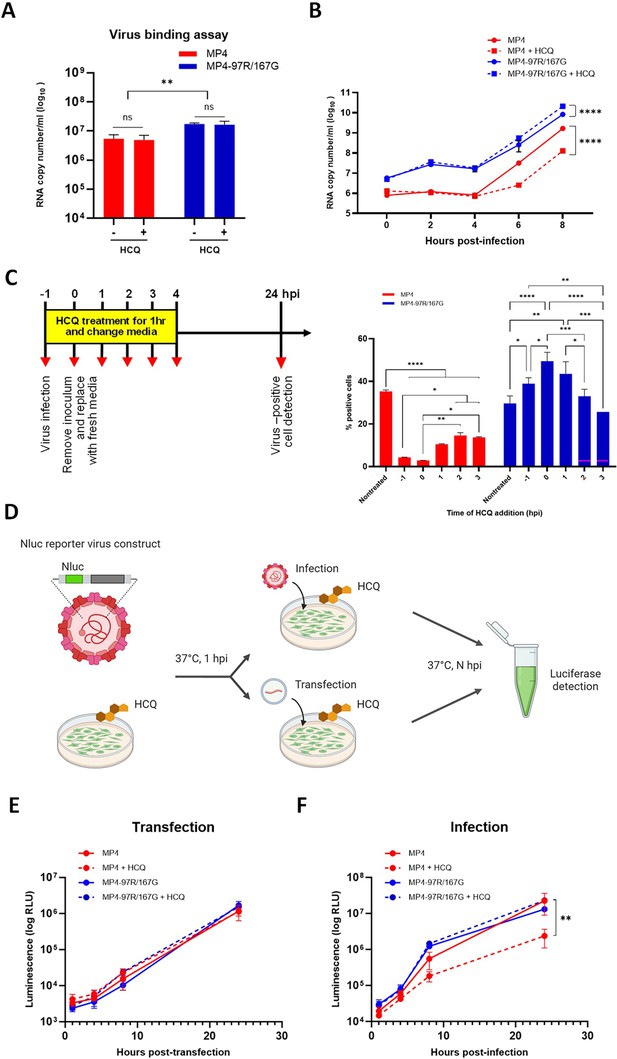
Hydroxychloroquine (HCQ) targets viral entry.
(A) Virus binding assay in Vero cells in presence of 25 µM HCQ. (B) Single-cycle replication kinetic in nontreated and HCQ-treated Vero cells. At each timepoint, cell lysates were collected, and viral RNA copy numbers were quantitated using RT-qPCR (C) Time-of-addition assay in Vero cells treated with HCQ starting at different timepoints. Infected cells (MOI 0.1) were quantitated 24 hpi by immunofluorescence (IF). (D) Schematic diagram of Vero cells pre-treated with HCQ and subsequently subjected to transfection of in vitro RNA transcript or infection with EV-A71 nanoluciferase (Nluc) reporter viruses. At the indicated timepoints, cell supernatants were collected, and luciferase activity was measured. Figure 2D was created with BioRender.com.(E & F) Results are expressed in % relative light unit (RLU) of treated versus nontreated virus at indicated timepoints. The mean and S.E.M from biological triplicates are shown. Statistical significance was calculated using two-way ANOVA, comparing treated and untreated control. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 2—source data 1
Related to Figure 2A.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig2-data1-v3.xlsx
-
Figure 2—source data 2
Related to Figure 2B.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig2-data2-v3.xlsx
-
Figure 2—source data 3
Related to Figure 2C.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig2-data3-v3.xlsx
-
Figure 2—source data 4
Related to Figure 2E.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig2-data4-v3.xlsx
-
Figure 2—source data 5
Related to Figure 2F.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig2-data5-v3.xlsx
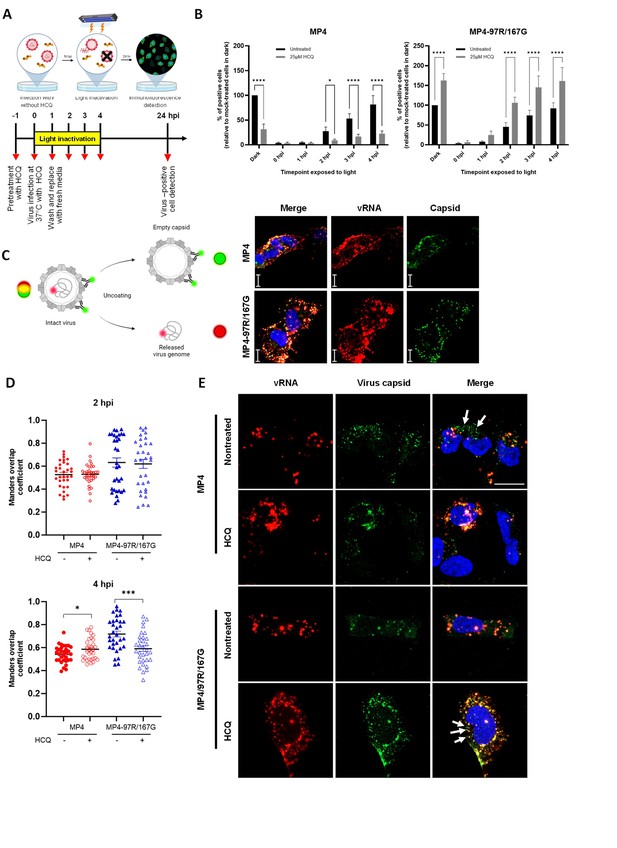
Hydroxychloroquine (HCQ) delays the uncoating of MP4.
(A) Schematic illustration of the neutral red assay workflow. Vero cells were pre-treated with or without HCQ for 1 hr. Neutral red-labeled viruses were allowed for cell infection at 37 °C for 1 hr (MOI 0.1). The inoculum was then removed and replaced with fresh media. Infected cells were exposed to light for 30 min at different timepoints and further incubated up to 24 hpi for immunofluorescence (IF) staining. Figure 3A was created with BioRender.com. (B) Effect of light inactivation on replication of neutral red-labeled MP4 (left panel) or MP4-97R/167G (right panel). Results are plotted as % of virus-positive cells relative to non-treated dark control. Mean and S.E.M of biological triplicates are shown. Statistical significances (two-way ANOVA) were calculated between treated and nontreated conditions. (C) Schematic illustration of virus uncoating monitored with the combinational use of RNA-FISH to detect EV-A71 RNA (red) and IF with anti-VP2 Ab to detect the viral capsid (green). Co-staining highlights intact viruses in yellow while empty capsids and free RNA are in green and red, respectively. (C, right panel) Representative images (scale, 20 µm) of MP4 and MP4-97R/167G binding after 1 hr at 4 °C with virus genomic RNA (vRNA) (red) and capsids (green). Figure 3C was created with BioRender.com. (D) Co-localization of capsid and vRNA in individual cells at 2 hpi and 4 hpi analyzed using Mander’s overlap coefficient (n=32 individual cells from two independent experiments). Statistical comparison (unpaired t-test) of untreated and treated groups. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (E) Representative images of the 4 hr time point. Arrows: empty capsid.
-
Figure 3—source data 1
Related to Figure 3B.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Related to Figure 3D.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig3-data2-v3.xlsx
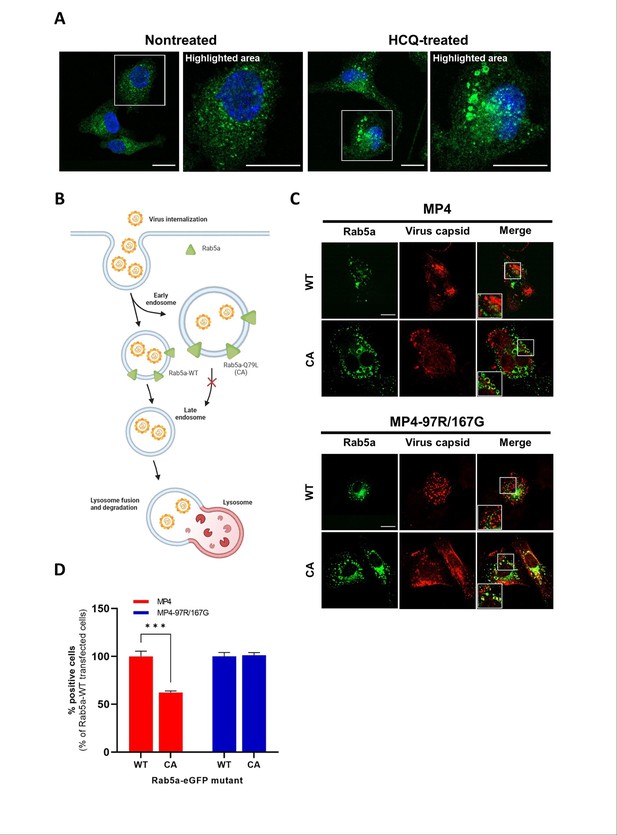
MP4-97R/167G uncoats from early endosomes.
(A) Nontreated and hydroxychloroquine (HCQ)-treated Vero cells were stained with anti-EEA-1 antibody (green) to label early endosomes, and DAPI (blue) to label cell nuclei. (B) Schematic representation of endosomal route upon overexpression of Rab5a WT or constitutively active (CA) mutant. In C and D, Vero cells transiently expressing Rab5a-eGFP WT or CA were fluorescence-activated flow cytometry (FACS)-sorted and infected with the two viral variants. Infections were compared at different time post-infection. Figure 4B was created with BioRender.com. In (C) viral capsids (anti-VP2 Ab, in red) localize in early endosomes at 2 hpi in cells expressing Rab5a WT or CA. In (D) the proportion of cells containing replicating viruses (stained with the anti-dsRNA J2 Ab, see Fig.S3B for representative images) is calculated at 7 hpi. Results and statistical significance (two-way ANOVA) are expressed relative to cells with Rab5a WT. Mean and S.E.M from triplicates are shown. ***p<0.001. In B and C, white boxes are enlarged in the right panel. Scale bar: 20 µm.
-
Figure 4—source data 1
Related to Figure 4D.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig4-data1-v3.xlsx
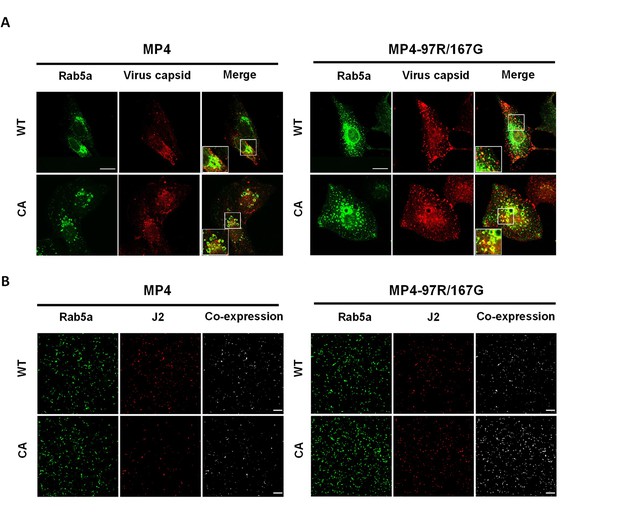
Localization of viral capsids and double-stranded RNA at respectively 0.5 hpi and 7 hpi in fluorescence-activated flow cytometry (FACS)-sorted GFP-positive Vero cells transiently expressing Rab5a-eGFP WT or constitutively active (CA) and infected with the two viral variants.
(A) Vero cells infected with MP4 and MP4-97R/167G were fixed at 0.5 hpi. Viral capsid localization was imaged with an anti-VP2 Ab (in red). Magnified areas are highlighted in white box and displayed at left bottom of merged image. Scale bar: 20 µm. (B) Vero cells infected with MP4 and MP4-97R/167G were fixed at 7 hpi and stained with anti-dsRNA J2 Ab to highlight viral replication. The images shown are representative examples of those used for the quantifications presented in Figure 4D. Scale bar: 400 µm.
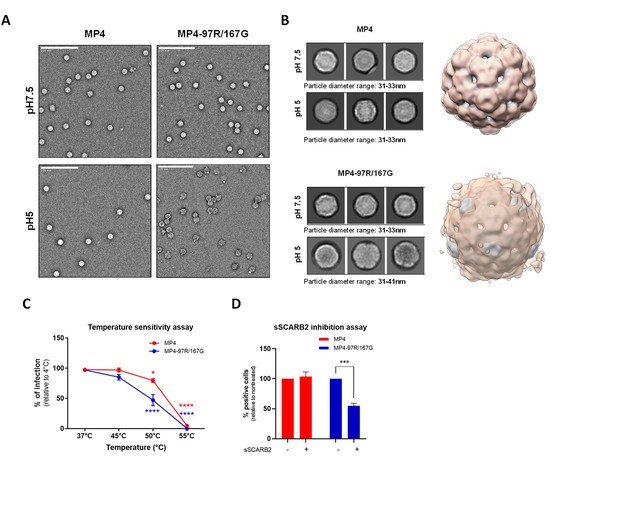
MP4 displayed stronger capsid stability and reduced sensitivity to acidification and high temperatures.
(A) Negative staining electron microscopy (nsEM) analysis of MP4 and MP4-97R/167G incubated at pH 7 and pH 5. Representative raw micrographs are shown in each case. (B) Representative 2D class averages generated from datasets shown in panel A (box size = 54 nm; left) and the overlay of the corresponding 3D maps (right). Gray and orange shade indicates virus particle reconstructions at pH 7 and pH 5, respectively. (C) Temperature sensitivity assay. Infected Vero cells (MOI 0.5) were quantitated by immunostaining with an anti-VP2 Ab at 24 hpi after 1 hr incubation at increasing temperatures. Results are shown as % of virus-positive cells relative to 4 °C treated control. Error bars indicate mean and S.E.M from biological triplicates. (D) For sSCARB2 inhibition assay, viruses (MOI 0.5) were incubated 1 hr at 37 °C with 1 µg of soluble scavenger receptor class B member 2 (SCARB2) (sSCARB2) before infection of Vero cells. Infected Vero cells were quantitated by immunostaining with an anti-VP2 Ab at 24 hpi. Results are shown as % of virus-positive cells relative to nontreated controls. Statistically significance was calculated with two-way ANOVA. ***p<0.001, ****p<0.0001.
-
Figure 5—source data 1
Related to Figure 5C.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig5-data1-v3.xlsx
-
Figure 5—source data 2
Related to Figure 5D.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig5-data2-v3.xlsx
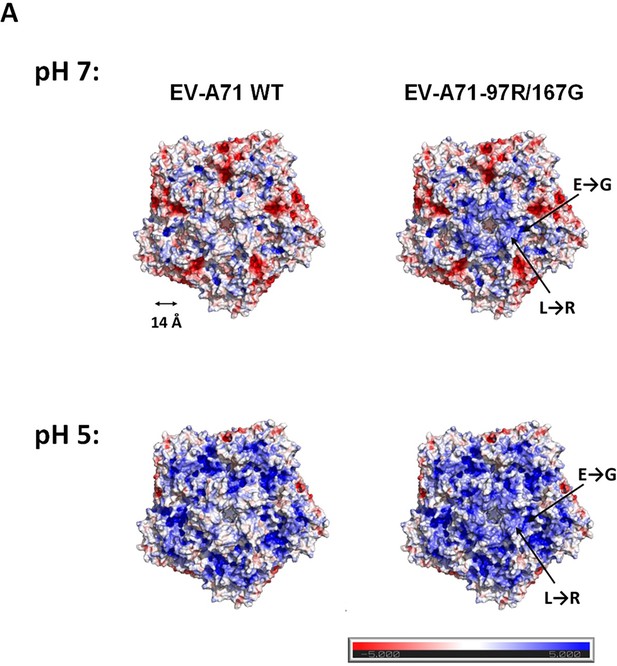
Visual presentation of prediction of changes induced by the L97R/E167G mutations on electrostatic surface potential.
Electrostatic surface potential calculated by Adaptive Poisson–Boltzmann Solver (APBS) from −5 kT/e (red) to +5 kT/e (blue).
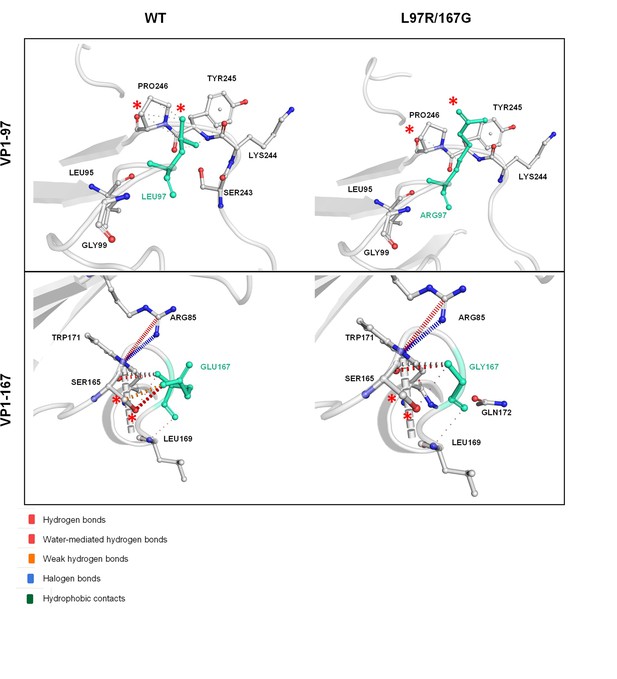
Prediction of changes in amino acid interactions and capsid stability performed using crystal structure of full assembled capsid on DynaMut server.
Interatomic interactions displayed and compared between WT and mutant capsid structures. VP1-97 and VP1-167 residues are labeled in light green and represented as sticks together with the surrounding interaction residues. Changes in interactions are highlighted on both WT and mutant structures with red asterisks (*).
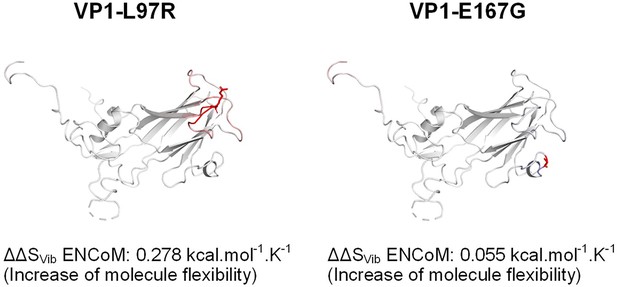
VP1-L97R and VP1-E167G mutations decrease capsid stability.
Computation of the vibrational entropy change (ΔΔSVib) between WT and mutants. Amino acids in red indicate an increase in molecule flexibility.
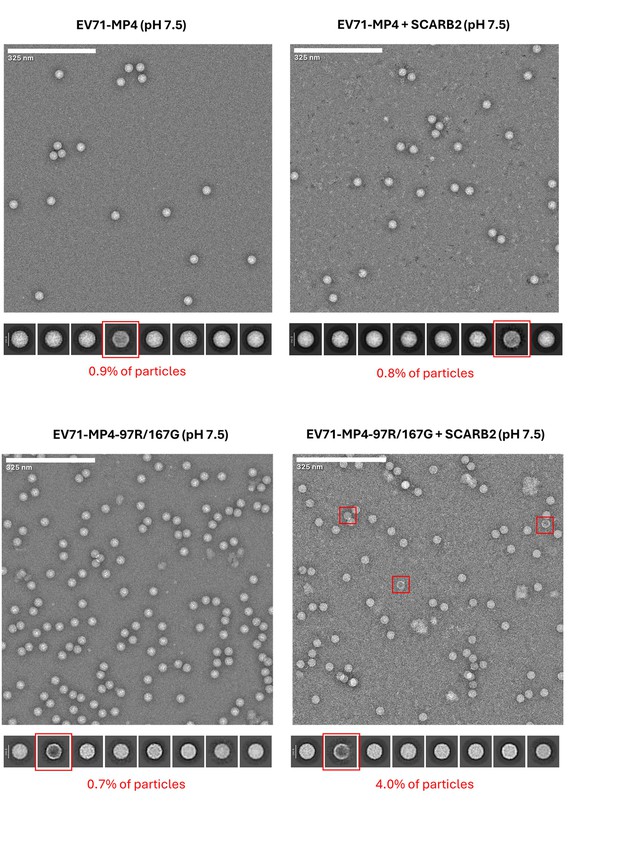
Raw micrographs and 2D classes of viruses incubated with soluble scavenger receptor class B member 2 (SCARB2).
Raw micrographs and 2D classes of MP4 and MP4-97R/167G incubated at neutral pH with and without sSCARB2. Representative 2D classes are shown in each case to recapitulate overall diversity in electron microscopy (EM) images (box size = 61 nm). Red squares indicate 2D class averages with strong staining in the middle of the particle indicating partially open capsids with low density of protein and genome components, which is consistent with empty capsids.
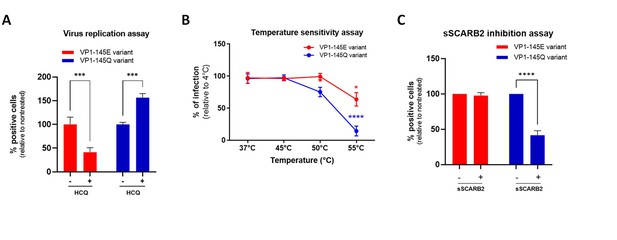
Heparan-sulfate-binding VP1-145Q variant exhibits resistance to hydroxychloroquine (HCQ) and higher sensitivity to soluble scavenger receptor class B member 2 (sSCARB2) inhibition and thermal stress.
(A) Virus inhibitory assay with VP1-145 variants were performed with 25 µg HCQ on Vero cells (MOI 0.1). (B) For temperature sensitivity assays, VP1-145 variants were incubated at increasing temperature for 1 hr before inoculated onto Vero cells (MOI 0.5). (C) For sSCARB2 inhibition assay, VP1-145 variants were incubated 1 hr at 37 °C with 1 µg of soluble SCARB2 (sSCARB2) before infection of Vero cells (MOI 0.5). Infected cells were quantitated by immunostaining with anti-VP2 Ab at 24 hpi. Results are shown as % of virus-positive cells relative to nontreated control (A & C) or 4 °C treated control (B). Mean and S.E.M of biological triplicates are shown. Statistically significant differences (two-way ANOVA) are shown. **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 6—source data 1
Related to Figure 6A.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig6-data1-v3.xlsx
-
Figure 6—source data 2
Related to Figure 6B.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig6-data2-v3.xlsx
-
Figure 6—source data 3
Related to Figure 6C.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig6-data3-v3.xlsx
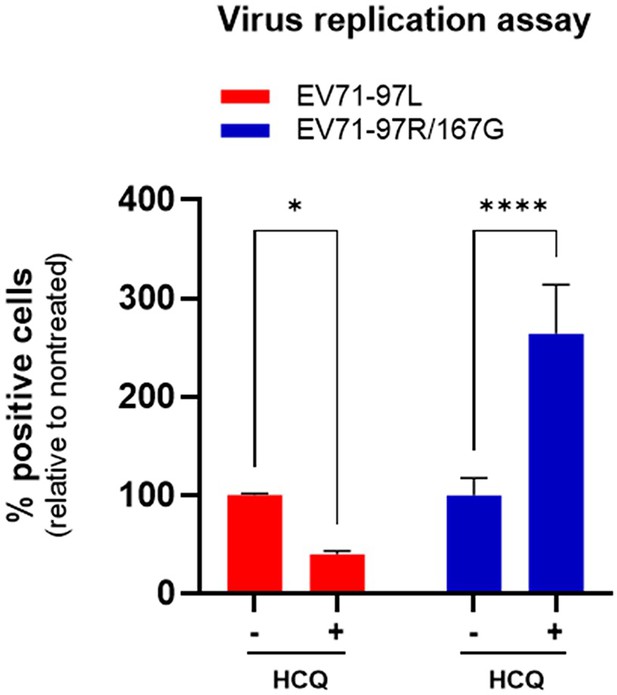
Human clinical strain variants exhibited similar hydroxychloroquine (HCQ) effects compared to MP4 variants.
Virus inhibitory assay with human clinical strain variant carries VP1-97L residue (EV71-97L) and VP1-97R/167G mutations (EV71-97R/167G) were treated with 25 µM HCQ on Vero cells (MOI 0.1). Infected cells were quantitated by immunostaining with anti-VP2 Ab at 24 hpi. Results are shown as % of virus-positive cells relative to nontreated control. Mean and S.E.M of biological triplicates are shown. Statistically significant differences (two-way ANOVA) are shown. *p<0.05, ****p<0.0001.
-
Figure 6—figure supplement 1—source data 1
Related to Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/98441/elife-98441-fig6-figsupp1-data1-v3.xlsx
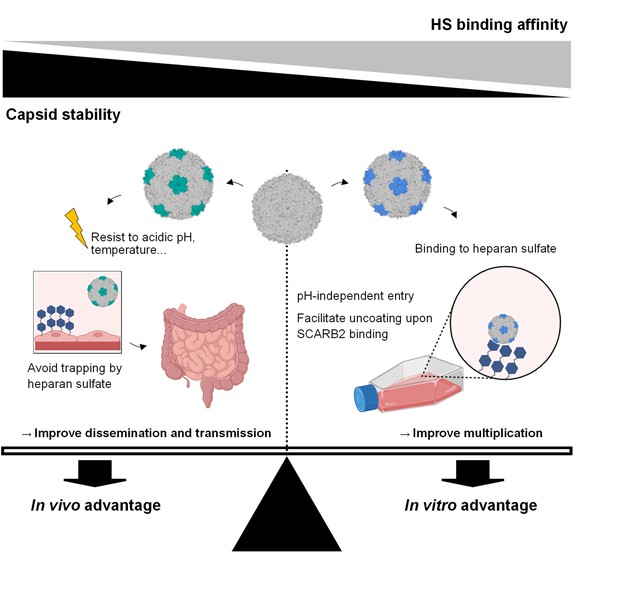
Seesaw model depicting the interplay between capsid mutations, heparan sulfate-binding, capsid stability as well as the resulting fitness changes in both in vitro and in vivo settings.
Viruses undergo continuous mutations to optimize fitness across diverse environments. In cell culture, they adapt to attain an ‘in vitro advantage’ by decreasing capsid stability while acquiring HS-binding capacity, consequently enhancing their infectivity. Conversely, during human infection, viruses adapt to secure an ‘in vivo advantage’ by bolstering capsid stability, relinquishing heparan sulfate (HS)-binding capacity, and thereby evading viral trapping and resisting environmental stresses. Figure 7 was created with BioRender.com.
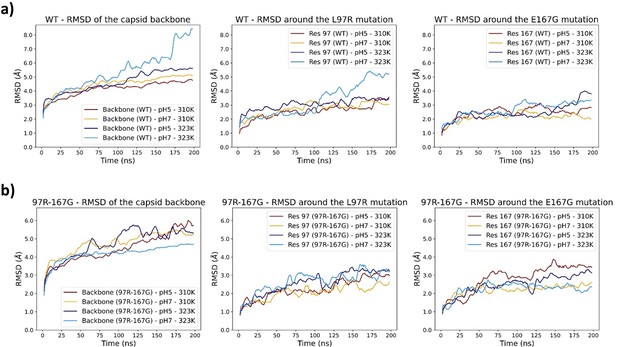
Molecular dynamic simulations of a subpart (corresponding to a capsid pentamer) of the wild type VP1 capsid (a) and the L97R-E167G mutated version (b), each at pH 5 and 7 and at 37°C (310K) and 50°C (323K).
The extraction of the subpart and the lack of physical constraints around the external-facing regions are inducing artificial moves during the simulation, and preventing its convergence, as witnessed by the high variability of root mean square deviations (RMSDs).
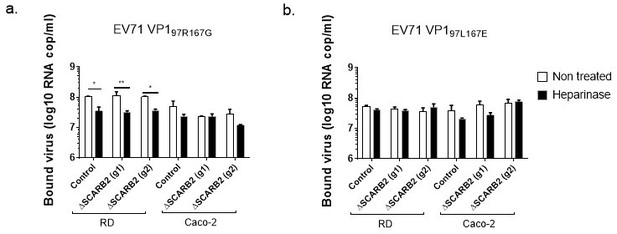
Binding of (a) EV71-VP1-97R167G and (b) EV71-VP-97L167E in RD and Caco-2 cells with and without SCARB2 (Cas9 CRISPR KO using two different guide RNAs, g1 and g2) and/or HS expression (heparinase treatment).
Viral loads were measured by RT-qPCR in whole cell extracts and are expressed as the mean ± SEM.
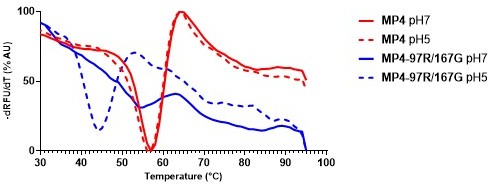
PaSTRy assay was performed by incubating virus in citrate phosphate buffer adjusted to pH7 and pH5.
Viral RNA release from capsid was detected using SYBR green II dye when the virus capsid was heating gradually with temperature increase of 1°C from 25°C to 95 °C.
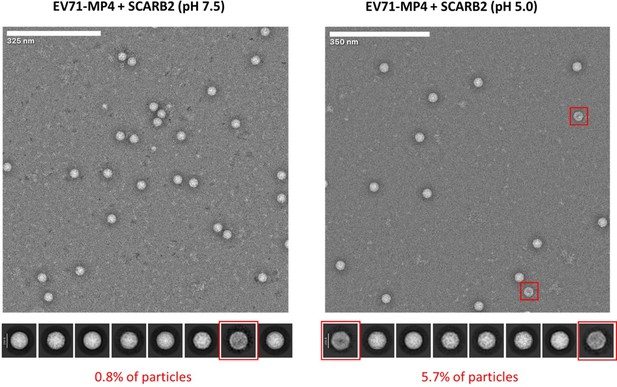
Raw micrographs and 2D classes of MP4 incubated with soluble SCARB2 for 30 minutes at neutral or acidic pH.
Red squares indicate 2D class averages with strong staining in the middle of the particle indicating partially open capsids with low density of protein and genome components, which is consistent with empty capsids.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-enterovirus 71 Ab (mouse monoclonal) | Merck Millipore | MAB979; RRID:AB_95300 | IF (1:1000) RNAscope (1:100) |
| Antibody | Anti-dsRNA monoclonal antibody J2 (mouse monoclonal) | Scicons | RRID:AB_2651015 | IF (1:500) |
| antibody | Anti-LAMP1 antibody (rabbit monoclonal) | Cell Signaling | D2D11, RRID:AB_2687579 | IF (1:100) |
| Antibody | Anti-EEA1 antibody (goat polyclonal) | Santa Cruz | sc-6414, RRID:AB_640035 | IF (1:100) |
| Antibody | Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (goat polyclonal) | Thermo Fisher Scientific | A-11029; RRID:A-11029 | IF (1:2000) RNAscope (1:200) |
| Antibody | Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (goat polyclonal) | Thermo Fisher Scientific | A-11032; RRID:AB_2534091 | IF (1:2000) |
| Strain, strain background (virus) | MP4 | Cagno et al., 2019 | GenBank: JN544419 | Mouse-adapted virus |
| Strain, strain background (virus) | HU-97L | Tee et al., 2021 | GenBank: EU414331 | Clinical strain |
| Strain, strain background (virus) | IEQ | Kobayashi and Koike, 2020 | GenBank: AF316321 | |
| Strain, strain background (virus) | IEE | Kobayashi and Koike, 2020 | GenBank: AF316321 | |
| Chemical compound, drug | Hydroxychloroquine | Tocris | 747-36-4 | |
| Chemical compound, drug | Bafilomycin A1 | InvivoGen | 88899-55-2 | |
| Chemical compound, drug | Sodium chlorate | Sigma | 7775-09-9 | |
| Chemical compound, drug | Neutral red | Sigma Aldrich | 553-24-2 | |
| Chemical compound, drug | Heparinase III | Amsbio | 37290-86-1 | |
| Chemical compound, drug | Puromycin | InvivoGen | 58-58-2 | |
| Chemical compound, drug | LysoTracker Deep Red | Thermo Fisher Scientific | LysoTracker Deep Red | |
| Chemical compound, drug | SYBR green II RNA gel stain | Thermo Fisher Scientific | SYBR green II RNA gel stain | |
| Recombinant protein | Recombinant Human LIMPII/SR-B2 Fc Chimera Protein, CF | Bio-Techne | 1966-LM | |
| Commercial kit or assay | RNAscope V-EV71-C1 probe | Biotechne | 1087481-C1 | |
| Commercial kit or assay | RNase P housekeeping gene | Thermo Fisher Scientific | 4316861 | |
| Commercial kit or assay | Lipofectamine 2000 | Thermo Fisher Scientific | 11668019 | |
| Commercial kit or assay | Superscript II reverse transcriptase | Thermo Fisher Scientific | 18064022 | |
| Commercial kit or assay | Random hexamer primers | Roche | 11034731001 | |
| Commercial kit or assay | Platinum Taq DNA Polymerase, DNA-free | Thermo Fisher Scientific | 15966025 | |
| Commercial kit or assay | TSA Vivid 570 kit | Tocris | 7526 | |
| Commercial kit or assay | E.Z.N.A. Viral RNA kit | Omega Bio-Tek | R6874-02 | |
| Commercial kit or assay | RNAscope Multiplex Fluorescent V2 | Biotechne | 323270 | |
| Commercial kit or assay | RNA-Protein Co-detection Ancillary Kit | Biotechne | 323180 | |
| Commercial kit or assay | KAPA SYBR FAST One-Step qRT-PCR Kits | Kapa Biosystems | KK4650 | |
| Commercial kit or assay | Nano-Glo Luciferase Assay System | Promega | N1110 | |
| Commercial kit or assay | Thiazolyl Blue Tetrazolium Bromide (MTT) | Merck | M5655 | |
| Commercial kit or assay | CyQUANT LDH Cytotoxicity Assay | Thermo Fisher Scientific | C20300 | |
| Cell line (Cercopithecus aethiops) | Vero cells | ATCC, USA | RRID:CVCL_0059 | |
| Cell line (Homo sapiens) | Rhabdomyosarcoma (RD) cells | ATCC, USA | RRID:CVCL_1649 | |
| Cell line (Homo sapiens) | RD-SCARB2-KO | Caroline Tapparel Yamayoshi et al., 2013 | ||
| Cell line (Homo sapiens) | RD-ΔEXT1+hSCARB2 | Satoshi Koike Kuronita et al., 2002 | ||
| Sequence-based reagent | RT-qPCR assay primer/for (Entero/Ge/08 assay) | Nishimura et al., 2024 | PCR primers | 5’-GCTGCGYTGGCGGCC-3’ |
| Sequence-based reagent | RT-qPCR assay primer/Rev (Entero/Ge/08 assay) | Nishimura et al., 2024 | PCR primers | 5’-GAAACACGGACACCCAAAGTAGT-3’ |
| Sequence-based reagent | RT-qPCR assay primer/probe (Entero/Ge/08 assay) | Nishimura et al., 2024 | PCR primers | 5’-CTCCGGCCCCTGAATGYGGCTAA-3’ |
| Recombinant DNA reagent | EV-A71/MP4 (Genbank accession number: JN544419; subgenogroup C2) | Jen-Reng Wang Guo et al., 2022 | ||
| Recombinant DNA reagent | IEQ (Genbank accession number: JN544419: AF316321; subgenogroup B4) | Jen-Reng Wang Kobayashi and Koike, 2020 | ||
| Recombinant DNA reagent | IEE (Genbank accession number: JN544419: AF316321; subgenogroup B4) | Jen-Reng Wang Kobayashi and Koike, 2020 | ||
| Recombinant DNA reagent | eGFP-Rab5a WT | Pierre-Yves Lozach Dang et al., 2014 | ||
| Recombinant DNA reagent | eGFP-Rab5a S34N | Pierre-Yves Lozach Dang et al., 2014 | ||
| Recombinant DNA reagent | eGFP-Rab5a Q79L | Pierre-Yves Lozach Dang et al., 2014 | ||
| Software | Geneious 10.2.16 | https://www.geneious.com | https://www.geneious.com | |
| Software | ImageXpress Micro XL | Molecular Devices | Molecular Devices | |
| Software | GraphPad Prism 9 | https://www.graphpad.com/scientific-software/prism/ | https://www.graphpad.com/scientific-software/prism/ | |
| Software | UCSF Chimera (version 1.13.1) | https://www.cgl.ucsf.edu/chimera/ | https://www.cgl.ucsf.edu/chimera/ |
Additional files
-
Supplementary file 1
Predicted Gibbs free energy change value (ΔΔG) was computed using I-mutant 2 server with calculation formula and an indication of protein structure stabilization as shown: Predicted Gibbs free energy change value (ΔΔG): ΔG (mutated protein) - ΔG (WT) in kcal/mol.
ΔΔG<0: Destabilizing mutation. ΔΔG>0: Stabilizing mutation.
- https://cdn.elifesciences.org/articles/98441/elife-98441-supp1-v3.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98441/elife-98441-mdarchecklist1-v3.pdf





