Identification of suitable target/E3 ligase pairs for PROTAC development using a rapamycin-induced proximity assay (RiPA)
Figures

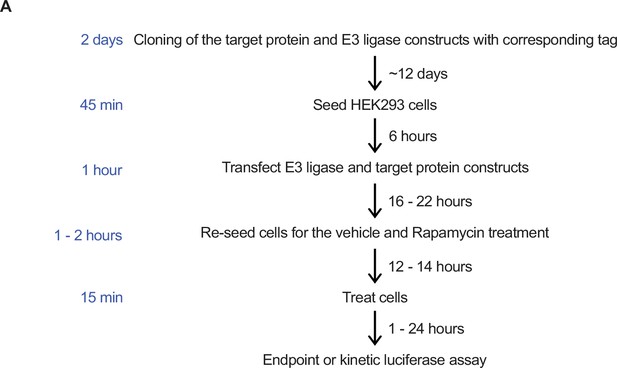
Rapamycin-induced proximity assay (RiPA) induces quantifiable degradation of target proteins.
(A) Schematic illustration of scenarios where proteolysis targeting chimera (PROTAC) could not induce degradation of a target protein. (B) Scheme of target protein (I) and E3 ligase or control (II) constructs used in RiPA. The linker indicated is 2x GSSG in all constructs unless stated otherwise. (C) Schematic describing the RiPA experimental protocol. (D) Immunoblot of WDR5 and VHL. HEK293 cells were co-transfected with WDR5-Luc-FKBP12 and VHL-FRB or FRB in the indicated ratio and treated with 100 nM rapamycin or vehicle for 6 hr after ~24 hr of expression. Vinculin was used as a loading control (as in all other immunoblotting experiments). (E) WDR5 levels based on luciferase measurements. HEK293 cells were co-transfected with WDR5-Luc-FKBP12 or Luc-WDR5-FKBP12 and VHL-FRB or FRB constructs in the indicated ratio, expressed for ~24 hr, and treated with rapamycin overnight. Bars represent mean ± s.d. of n=3 replicates. (F) Immunoblot of WDR5 and VHL. HEK293 cells were co-transfected with a combination of WDR5-Luc-FKBP12 or Luc-WDR5-FKBP12 and VHL-FRB or FRB-VHL or FRB in the ratio of 1:10, expressed for ~24 hr and treated with rapamycin overnight. WDR5 and VHL fusion proteins tagged at the N- and C-terminal show different migration behaviors despite having same molecular weight.
-
Figure 1—source data 1
PDF file containing original western blot for Figure 1D and F, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig1-data1-v1.zip
-
Figure 1—source data 2
Original files for western blot analysis displayed in Figure 1D and F.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig1-data2-v1.zip
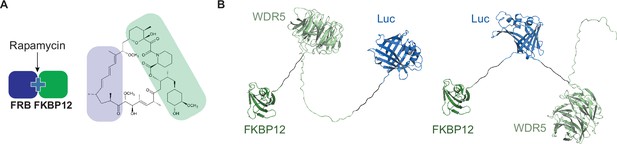
Rapamycin-induced proximity assay (RiPA) induces quantifiable degradation of target proteins.
(A) Schematic illustration of rapamycin-induced dimerization of FRB and FKBP12 and structure of rapamycin. (B) Structure of WDR5-Luc-FKBP12 and Luc-WDR5-FKBP12 fusion proteins. A flexible linker, 2x GSSG is present between each component.

Rapamycin-induced proximity assay (RiPA) correctly predicts suitability of E3 ligases for WDR5 proteolysis targeting chimeras (PROTACs).
(A) Immunoblot of WDR5, VHL, and CRBN. HEK293 cells were transfected with WDR5-Luc-FKBP12 and VHL-FRB or CRBN-FRB or FRB in a 1:10 ratio, expressed for 24 hr, and treated with 10 nM rapamycin for 6 hr. (B) WDR5 levels based on luciferase measurement. Luminescence was measured in HEK293 cells as described in (A) after 10 nM rapamycin treatment at specified time points. Data are shown as individual values (n=3 replicates), with a curve fitted to the mean across replicates. (C) Immunoblot of AURKA, VHL, and CRBN. HEK293 cells were transfected with AURKA-Luc-FKBP12 and VHL-FRB or CRBN-FRB or FRB in a 1:10 ratio and treated with 10 nM rapamycin for 6 hr. (D) AURKA levels based on luciferase measurement. Luminescence was measured in HEK293 cells as described in (C) after 10 nM rapamycin treatment at indicated time points. Data are shown as individual values (n=3 replicates), with a curve fitted to the mean across replicates. (E) Structure of FKBP12 and luciferase. Molecular surface representation of FKBP12 (top) and luciferase (bottom) showing lysine residues on their surface. The lysine residues are labeled and two sides for each protein are shown. (F) WDR5 levels based on luciferase measurement. HEK293 cells were co-transfected with either WDR5-Luc-FKBP12 (WT) or WDR5-Luc-FKBP12 construct where all lysine residues on Luc and FKBP12 were mutated to arginine (Kless) and FRB, expressed for ~24 hr and luminescence measured. Bars represent mean ± s.d. of n=12 replicates. (G) WDR5 levels based on luciferase measurement. Luminescence was measured in HEK293 cells expressing WDR5-Luc-FKBP12(Kless) and VHL-FRB or CRBN-FRB or FRB after 10 nM rapamycin treatment at specified time points. Data are shown as individual values (n=3 replicates), with a curve fitted to the mean across replicates. (H) KRASG12D levels based on luciferase measurements. HEK293 cells were co-transfected with KRASG12D-Luc-FKBP12(Kless) and VHL-FRB or FRB constructs, expressed for ~24 hr, and treated with 10 nM rapamycin (Rapa.) in the presence or absence of 10 µM MG132 and 5 µM MLN4924 for 8 hr. Bars represent mean ± s.d. of n=2 replicates.
-
Figure 2—source data 1
PDF file containing original western blot for Figure 2A and C, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig2-data1-v1.zip
-
Figure 2—source data 2
Original files for western blot analysis displayed in Figure 2A and C.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig2-data2-v1.zip
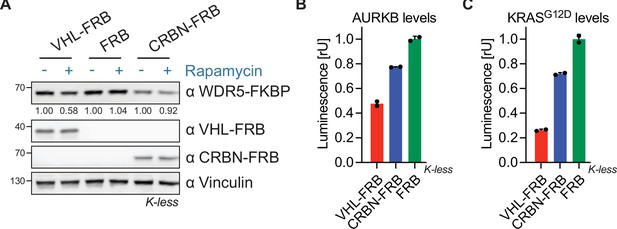
Rapamycin-induced proximity assay (RiPA) predicts suitability of E3 ligases for proteolysis targeting chimeras (PROTACs) against various targets.
(A) Immunoblot of WDR5, VHL, and CRBN. HEK293 cells expressing WDR5-Luc-FKBP12(Kless) and VHL-FRB or CRBN-FRB or FRB were treated with 10 nM rapamycin for 6 hr. (B) AURKB levels based on luciferase measurement. Luminescence was measured in HEK293 cells expressing AURKB-Luc-FKBP12(Kless) and VHL-FRB or CRBN-FRB or FRB after 10 nM rapamycin treatment for 8 hr. Data represent mean ± s.d. of n=2 replicates. (C) KRASG12D levels based on luciferase measurement. Luminescence was measured in HEK293 cells expressing KRASG12D-Luc-FKBP12(Kless) and VHL-FRB or CRBN-FRB or FRB after 10 nM rapamycin treatment for 8 hr. Data represent mean ± s.d. of n=2 replicates.
-
Figure 2—figure supplement 1—source data 1
PDF file containing original western blot for Figure 1A, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Original files for western blot analysis displayed in Figure 1A.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig2-figsupp1-data2-v1.zip
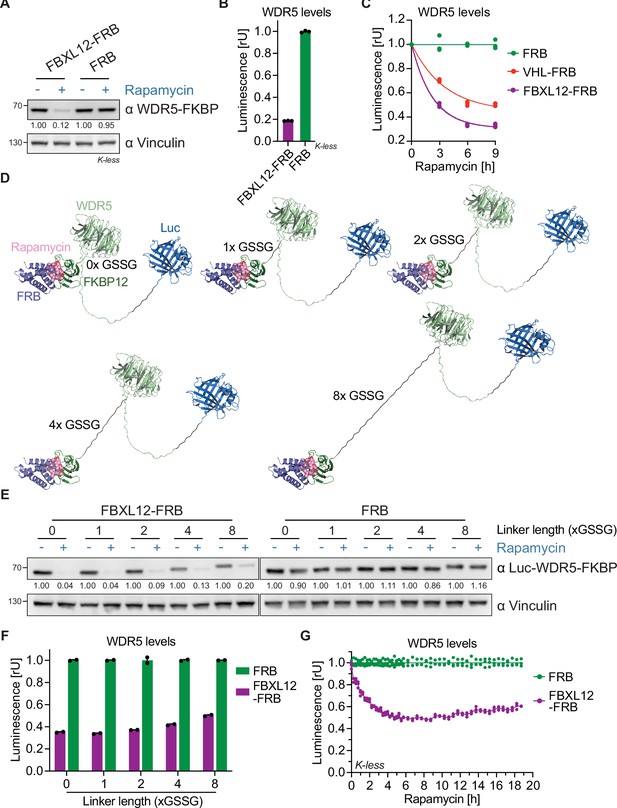
Rapamycin-induced proximity assay (RiPA) can identify degradative E3 ligases not previously used for proteolysis targeting chimeras (PROTACs).
(A) Immunoblot of WDR5. HEK293 cells were transfected with WDR5-Luc-FKBP12(Kless) and FBXL12-FRB or FRB in a ratio of 1:10 and treated with 10 nM rapamycin for 8 hr. (B) WDR5 levels based on luciferase measurement. Luminescence of WDR5-Luc-FKBP12(Kless) in the same cells as in (A). Bars represent mean ± s.d. of n=3 replicates. (C) WDR5 levels based on luciferase measurement. HEK293 cells were transfected with WDR5-Luc-FKBP12(Kless) and FBXL12-FRB or VHL-FRB or FRB in a ratio of 1:1000 and treated with 10 nM rapamycin for the indicated time point. Data are shown as individual values (n=3 replicates), with a curve fitted to the mean across replicates. (D) Model of Luc-WDR5-FKBP12 constructs. Structure of Luc-WDR5-FKBP12 with indicated linkers between WDR5 and FKBP12 bound to rapamycin and FRB. The linker between Luc-WDR5 is always 2x GSSG. (E) Immunoblot of WDR5. HEK293 cells were transfected with Luc-WDR5-FKBP12 containing indicated linker length and FBXL12-FRB or FRB in the ratio of 1:100, expressed for ~24 hr, and treated with 10 nM rapamycin for 6 hr. (F) WDR5 levels based on luciferase measurement. Luminescence of Luc-WDR5-FKBP12 constructs as in cells from (E) and treated with 10 nM rapamycin for 8 hr. Bars represent mean ± s.d. of n=2 replicates. (G) WDR5 levels based on kinetic luciferase measurement. HEK293 cells were transfected with WDR5-Luc-FKBP12(Kless) and FBXL12-FRB or FRB in a ratio of 1:100, expressed for ~24 hr, treated with 10 nM rapamycin, and luminescence measured for 19 hr. Data are shown as individual values (n=3 replicates).
-
Figure 3—source data 1
PDF file containing original western blot for Figure 3A and E, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig3-data1-v1.zip
-
Figure 3—source data 2
Original files for western blot analysis displayed in Figure 3A and E.
- https://cdn.elifesciences.org/articles/98450/elife-98450-fig3-data2-v1.zip
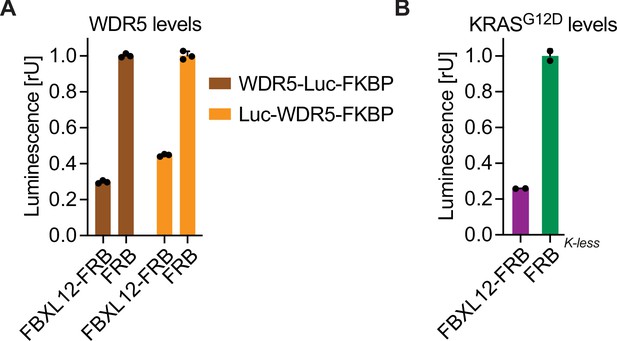
Rapamycin-induced proximity assay (RiPA) can identify degradative E3 ligases not previously used for proteolysis targeting chimeras (PROTACs).
(A) WDR5 levels based on luciferase measurement. HEK293 cells were transfected with Luc-WDR5-FKBP12 or WDR5-Luc-FKBP12 and FBXL12-FRB or FRB in a ratio of 1:10, expressed for ~24 hr, and treated with 10 nM rapamycin for 9 hr. Bars represent mean ± s.d. from n=3 replicates. (B) KRASG12D levels based on luciferase measurements. HEK293 cells were co-transfected with KRASG12D-Luc-FKBP12(Kless) and FBXL12-FRB or FRB constructs, expressed for ~24 hr, and treated with 10 nM rapamycin for 8 hr. Bars represent mean ± s.d. of n=2 replicates.

Identification of degradation-inducing E3 ligases by designing a universal substrate.
(A) Model of lysine-rich luciferase. Structure of mutated luciferase with 5 additional (LucV1) and 12 additional (LucV2) lysines as compared to 7 lysine residues of wild-type luciferase. The lysine residues from WT (black), LucV1 (red), and Luc V2 (orange; red as in V1) are labeled. (B) Scheme of wild-type luciferase (WT) or lysine-rich luciferase (V1, V2, K3, K6, and K12) containing constructs. (C) Luciferase measurement. HEK293 cells were co-transfected with Luc-FKBP12 constructs as shown in (B) and FRB, expressed for ~24 hr, and luminescence was compared. Bars represent mean ± s.d. of n=2 replicates. (D) Luciferase measurement. HEK293 cells were transfected with the indicated versions of Luc-FKBP12 and FBXL12-FRB or FRB in a ratio of 1:100, expressed for ~24 hr, and treated with 10 nM rapamycin for 8 hr. Bars represent mean ± s.d. of n=2 replicates. (E) Kinetic luminescence measurement. HEK293 cells expressing constructs as described in (D) were treated with 10 nM rapamycin or vehicle and luminescence was monitored for 22 hr. Data are shown as individual values (n=2 replicates).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-WDR5 (G-9) (mouse monoclonal) | Santa Cruz Biotechnology | RRID: AB_3331659 | WB (1:1000) |
| Antibody | anti-AuroraA/AIK (rabbit, polyclonal) | Cell Signaling Technology | RRID: AB_2061342 | WB (1:1000) |
| Antibody | anti-VHL (VHL40) (mouse monoclonal) | Santa Cruz Biotechnology | RRID: AB_2215955 | WB (1:1000) |
| Antibody | anti-CRBN (D8H3S) (rabbit, monoclonal) | Cell Signaling Technology | RRID: AB_2799810 | WB (1:1000) |
| Antibody | anti-vinculin (mouse monoclonal) | Sigma-Aldrich | RRID: AB_477629 | WB (1:2000) |
| Antibody | ECL-Anti-rabbit IgG Horseradish Peroxidase | GE Healthcare | Cat# NA934V | WB (1:7500) |
| Antibody | ECL-Anti-mouse IgG Horseradish Peroxidase | GE Healthcare | Cat# NA931V | WB (1:7500) |
| Chemical compound, drug | Rapamycin | Selleckchem | Cat# S1039 | |
| Chemical compound, drug | MG132 | Calbiochem/Merck | Cat# 474790 | |
| Chemical compound, drug | Pevonedistat (MLN4924) | Selleckchem | Cat# S7109 | |
| Chemical compound, drug | Protease Inhibitor Cocktail | Sigma-Aldrich | Cat# P8340 | |
| Chemical compound, drug | Phosphatase Inhibitor Cocktail 2 | Sigma-Aldrich | Cat# P5726 | |
| Chemical compound, drug | Phosphatase Inhibitor Cocktail 3 | Sigma-Aldrich | Cat# P0044 | |
| Peptide, recombinant protein | Phusion High-Fidelity DNA Polymerase | Thermo Fisher Scientific | Cat# F530L | |
| Chemical compound, drug | Immobilon Western Chemiluminescent HRP Substrate | Merck Millipore | Cat#WBKLS0500 | |
| Peptide, recombinant protein | NanoGlo Endurazine Live Cell Substrate | Promega | Cat# N2571 | |
| Peptide, recombinant protein | Phusion Plus DNA Polymerase | Thermo Fisher Scientific | Cat# F630L | |
| Peptide, recombinant protein | AgeI-HF | New England BioLabs | Cat# R3552L | |
| Peptide, recombinant protein | AscI | New England BioLabs | Cat# R0558L | |
| Peptide, recombinant protein | BamHI-HF | New England BioLabs | Cat# R3136L | |
| Peptide, recombinant protein | EcoRI-HF | New England BioLabs | Cat# R3101L | |
| Peptide, recombinant protein | MluI-HF | New England BioLabs | Cat# R3198L | |
| Peptide, recombinant protein | SpeI-HF | New England BioLabs | Cat# R3133L | |
| Peptide, recombinant protein | XhoI | New England BioLabs | Cat# R0146L | |
| Commercial assay or kit | Nano-Glo Luciferase Assay | Promega | Cat# N1120 | |
| Commercial assay or kit | GeneJET Gel Extraction Kit | Thermo Fisher Scientific | Cat# K0692 | |
| Commercial assay or kit | PureLink HiPure Plasmid Maxiprep Kit | Invitrogen | Cat# K210007 | |
| Cell line (Homo-sapiens) | human embryonic kidney 293T | ATCC | CRL-3216 | |
| Recombinant DNA reagent | pRRL_puro (plasmid) | PMID:25043018 | ||
| Recombinant DNA reagent | pRRL_hygro (plasmid) | PMID:25043018 | ||
| Recombinant DNA reagent | pRRL_puro_WDR5-Luc-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc-WDR5-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_hygro_FRB-VHL (plasmid) | This paper | See Materials and methods, Section ‘Cloning of E3-FRB constructs’ | |
| Recombinant DNA reagent | pRRL_hygro_VHL-FRB (plasmid) | This paper | See Materials and methods, Section ‘Cloning of E3-FRB constructs’ | |
| Recombinant DNA reagent | pRRL_hygro_FRB (plasmid) | This paper | See Materials and methods, Section ‘Cloning of E3-FRB constructs’ | |
| Recombinant DNA reagent | pRRL_hygro_CRBN-FRB (plasmid) | This paper | See Materials and methods, Section ‘Cloning of E3-FRB constructs’ | |
| Recombinant DNA reagent | pRRL_puro_AURKA-Luc-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_WDR5-Luc-FKBP12(Kless) (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_AURKB-Luc-FKBP12(Kless) (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_KRASG12D-Luc-FKBP12(Kless) (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_hygro_FBXL12-FRB (plasmid) | This paper | See Materials and methods, Section ‘Cloning of E3-FRB constructs’ | |
| Recombinant DNA reagent | pRRL_puro_C-term_Luc-FKBP12_EV (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_C-term_Luc-FKBP12(Kless)_EV (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_hygro_E3-FRB_EV (plasmid) | This paper | See Materials and methods, Section ‘Cloning of E3-FRB constructs’ | |
| Recombinant DNA reagent | pRRL_hygro_FRB-E3_EV (plasmid) | This paper | See Materials and methods, Section ‘Cloning of E3-FRB constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc-WDR5-FKBP12_0xGSSG (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc-WDR5-FKBP12_1xGSSG (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc-WDR5-FKBP12_2xGSSG (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc-WDR5-FKBP12_4xGSSG (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc-WDR5-FKBP12_8xGSSG (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Target-FKBP12 constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Luc-FKBP12 universal substrate constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc_V1-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Luc-FKBP12 universal substrate constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc_V2-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Luc-FKBP12 universal substrate constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc_K3-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Luc-FKBP12 universal substrate constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc_K6-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Luc-FKBP12 universal substrate constructs’ | |
| Recombinant DNA reagent | pRRL_puro_Luc_K12-FKBP12 (plasmid) | This paper | See Materials and methods, Section ‘Cloning of Luc-FKBP12 universal substrate constructs’ | |
| Sequence-based reagent | Oligonucleotides used for PCR and cloning | Sigma-Aldrich | See Supplementary file 1 | |
| Sequence-based reagent | IDT G-blocks (double stranded linear DNA) | Integrated DNA technologies | See Supplementary file 2 | |
| Software, algorithm | GraphPad PRISM 10.1.1 | GraphPad | https://www.graphpad.com/ | |
| Software, algorithm | ImageJ 1.52 | PMID:22930834 | https://imagej.nih.gov/ij/ | |
| Software, algorithm | PyMOL Molecular Graphics System | Schrödinger, LLC | https://pymol.org/ |
Additional files
-
Supplementary file 1
Amino acid sequences.
The table contains all the amino acid sequences of the constructs used in this study.
- https://cdn.elifesciences.org/articles/98450/elife-98450-supp1-v1.xlsx
-
Supplementary file 2
Oligonucleotide sequences.
The table contains all oligonucleotide sequences, primers, and gBlocks used in this study.
- https://cdn.elifesciences.org/articles/98450/elife-98450-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98450/elife-98450-mdarchecklist1-v1.pdf