Single turnover transient state kinetics reveals processive protein unfolding catalyzed by Escherichia coli ClpB
Figures
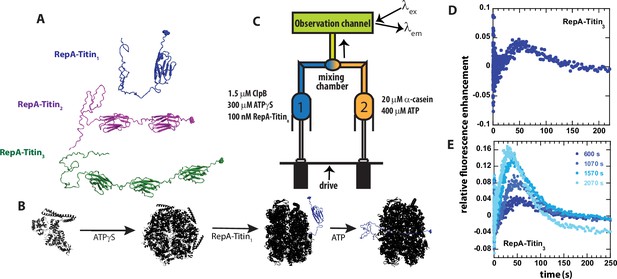
Single turnover ClpB catalyzed protein unfolding.
(A) RepA(1-70)-Titin1 (Blue), RepA(1-70)-Titin2 (purple), and RepA(1-70)-Titin3 (green). Each construct from N- to C-terminus consists of the first 70 amino acids of the Phage P1 RepA protein, a known binding sequence for ClpB followed by tandem repeats of the Titin I27 domain separated by linkers. Each construct contains a single cysteine shown in space-filling at the C-terminus that has been reacted with Alex Fluor (AF)–555. (B) Schematic of steps in forming pre-bound complex based on our previous work (Lin and Lucius, 2015; Lin and Lucius, 2016; Weaver et al., 2017; Li et al., 2015a). ClpB (black) is assembled into hexameric rings competent for substrate binding by adding ATPγS, illustrated as bound to the RepA-Titin1 substrate in blue, followed by rapid mixing with ATP. As shown, ClpB is expected to unfold the Titin I27 domains and translocate the newly unfolded substrates through the axial channel of the hexameric ring. (C) Schematic representation of stopped-flow. Syringe 1 contains the indicated concentrations of ClpB monomer, ATPγS, and RepA-TitinX, where X = 1, 2, 3. Syringe 2 contains 400 μM ATP and 20 μM α-casein to serve as a trap for any free ClpB. The contents of the two syringes are rapidly mixed at a 1:1 mixing ratio and flow into the observation channel where AF555 is excited at λex = 555 nm and emission is observed at λem > 570 nm. (D) Representative time-course collected using strategy in (C) using RepA-Titin3 after pre-incubating the sample at 25°C for 600 s. (E) Successive experimental time-courses collected as in D. The total time of incubation before collection of the time-course is indicated.
-
Figure 1—source data 1
The data points for relative fluorescence enhancement of Figure 1D, E are tabulated as a function of time for each incubation period of 600, 1070, 1570, and 2070 s.
Please note that Figure 1D and E have the exact same data for an incubation time of 600 s.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig1-data1-v1.xlsx
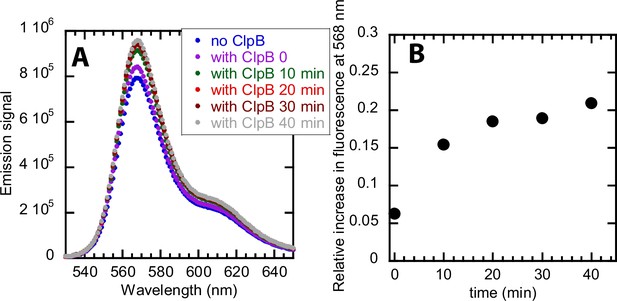
Testing protein-induced fluorescence enhancement with an unstructured substrate of 50 amino acids.
(A) 2 μM ClpB in the presence of 1 mM ATPγS was incubated with 0.5 μM polypeptide labeled with AF555. The emission spectra by exciting AF555 at 520 nm were collected every 10 min. The blue trace represents emission spectra collected with no ClpB. The purple trace represents emission spectra collected immediately after mixing ClpB with ATPγS and polypeptide. 10 min (green), 20 min (red), 30 min (brown), and 40 min (gray) represent the incubation times of ClpB with ATPγS and polypeptide. (B) Relative fluorescence enhancement was calculated at 568 nm by using Equation 4 and is plotted as a function of incubation times of ClpB with ATPγS and polypeptide. With the increase in incubation times, we observe fluorescence enhancement upon binding of ClpB with polypeptide substrate.
-
Figure 1—figure supplement 1—source data 1
The sheet labeled ‘Figure 1—figure supplement 1A’ consists of data points for emission signal as a function of wavelength shown in Figure 1—figure supplement 1A.
The column labeled ‘no ClpB’ is the data under no ClpB conditions. The columns with ‘+ClpB’ and ‘0, 10, 20, 30, 40 min’ represent the data for conditions with ClpB at different incubation times. The sheet labeled ‘Figure 1—figure supplement 1B’ tabulates the data points for Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig1-figsupp1-data1-v1.xlsx
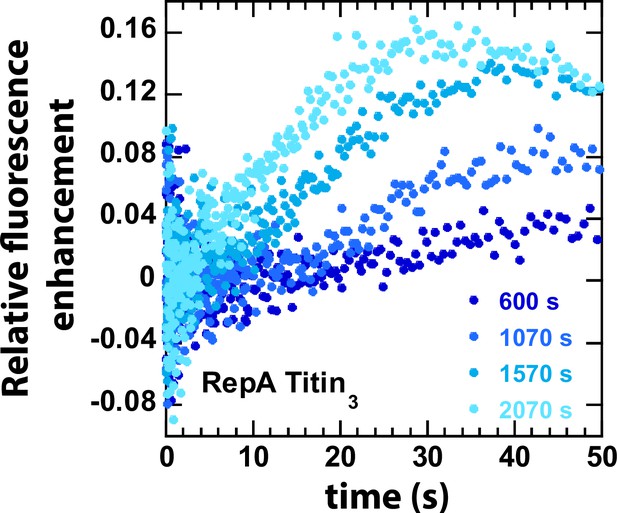
A zoomed-in version of Figure 1E to show the decreasing lag with increasing incubation time of the pre-bound complex.
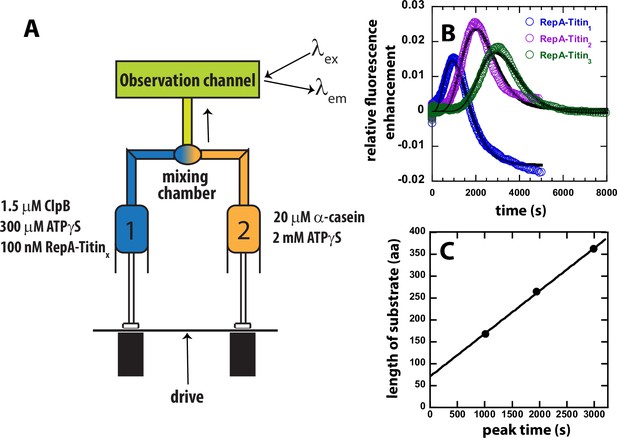
Test for ATPγS-driven protein unfolding by ClpB.
(A) Mixing strategy as in Figure 1C but with ATP replaced with 2 mM ATPγS in Syringe 2. (B) Time-courses collected using RepA-Titin1 (blue), RepA-Titin2 (purple), and RepA-Titin3 (green) plotted as relative fluorescence enhancement vs. time. The solid black line represents the best-fit line from fitting to Scheme 1, Figure 4A. The fitting parameters obtained are the unfolding rate constant, kU = (0.0042 ± 0.0003) s–1, and the kinetic step-size, m = (26 ± 5) aa. (C) Length of substrate vs. peak time determined from B. The plot was fit to a linear equation to yield a slope = (0.098 ± 0.003) aa s–1 and intercept of (71 ± 7) aa.
-
Figure 2—source data 1
The sheet labeled ‘Figure 2B' consists of data points in a tabulated form for each RepA-TitinX substrate, as shown in Figure 2B.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig2-data1-v1.xlsx
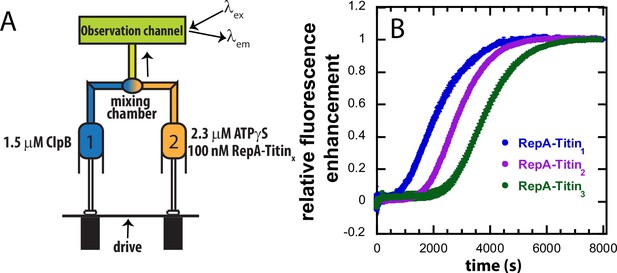
Post-mix assay to investigate ATPγS driven protein unfolding catalyzed by ClpB.
(A) 1.5 μM ClpB (Syringe 1) is mixed with 2.3 μM ATPγS (Syringe 2) and 100 nM RepA-TitinX (Syringe 2), where X = 1, 2, 3, using standard mix on stopped-flow apparatus. Under these conditions, upon mixing, ClpB binds ATPγS, assembles into hexamers competent for binding RepA-Titin1 and proceeds to unfolding. In this setup, the fluorescence change is monitored over time. (B) The scatter plot shows the experimental time-courses with the maximum fluorescence normalized to one obtained using three different RepA-TitinX substrates. The experimental time-courses are an average of at least three or more successive time-courses and the standard deviation of these averaged time-courses are shown as error bars on the data points.
-
Figure 2—figure supplement 1—source data 1
The sheet consists of data points in a tabulated form for each RepA-TitinX substrate, as shown in Figure 2—figure supplement 1. The error bars for each relative fluorescence enhancement data point are tabulated here as standard deviation.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig2-figsupp1-data1-v1.xlsx
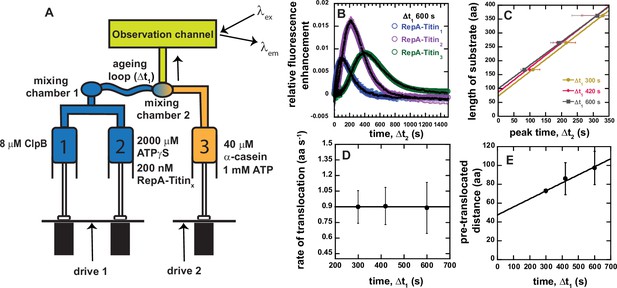
Sequential mixing stopped-flow strategy.
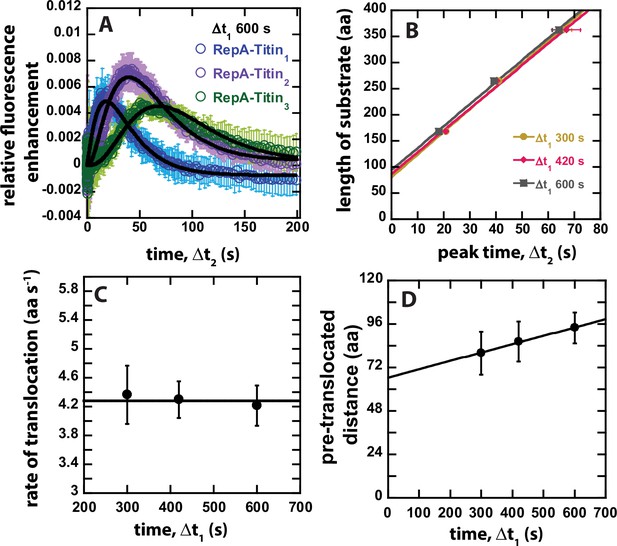
(A) Schematic representation of sequential mixing. Syringe 1 contains 8 μM ClpB monomer, Syringe 2 contains 2 mM ATPγS and 200 nM RepA-TitinX, and Syringe 3 contains 40 μM α-casein and 1 mM ATP. The contents of Syringes 1 and 2 are mixed 1:1 in mixing chamber 1 leading to a concentration of 4 μM ClpB, 1 mM ATPγS, and 100 nM RepA-TitinX. The sample ages for a user-defined amount of time, Δt1, followed by rapid mixing with the contents of Syringe 3. The sample flows into the observation channel at a final concentration of 2 μM ClpB, 50 nM RepA-TitinX, 500 μM ATPγS, 500 μM ATP, and 20 μM α-casein. In the observation channel AF555 is excited at λex = 555 nm and emission is observed at λem > 570 nm. (B) Representative time-courses from the average of five or more sequentially collected time-courses for RepA-Titin1(blue), RepA-Titin2(purple), and RepA-Titin3(green) at Δt1 = 600 s. The black solid lines represent the best-fit line from fitting to Scheme 1 (see Figure 4A). The fitting parameters obtained are kU = (0.017 ± 0.002) s−1 and m = (56.5 ± 0.7) aa. (C) Total length of substrate as a function of peak time determined for Δt1 = 300, 420, and 600 s. Solid lines represent weighted linear fits yielding (D) slope vs. Δt1 and (E) the intercept vs. Δt1. All data points and error bars represent the average and standard deviation determined from three replicates.
-
Figure 3—source data 1
The sheet labeled as ‘Figure 3B' consists of data points in a tabulated form for each RepA-TitinX substrate, as shown in Figure 3B.
The error bars for each relative fluorescence enhancement data point are tabulated here as standard deviation. The data points corresponding to the fits shown in black in Figure 3B are tabulated after ‘fit data to Scheme 1' for each RepA-TitinX substrate. The sheet labeled ‘Figure 3C' are data points for the total length vs. peak time plot shown in Figure 3C at each Δt1. The sheet labeled ‘Figure 3D' has data points for the rate of translocation as a function of Δt1 shown in Figure 3D. The error bars represented in Figure 3D are tabulated as the standard deviation in this sheet for each corresponding rate of translocation. The sheet labeled ‘Figure 3E’ consists of data points for pre-translocated distance as a function of Δt1, as shown in Figure 3E. The error bars represented in Figure 3E are tabulated as the standard deviation in this sheet for each corresponding pre-translocated distance.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig3-data1-v1.xlsx
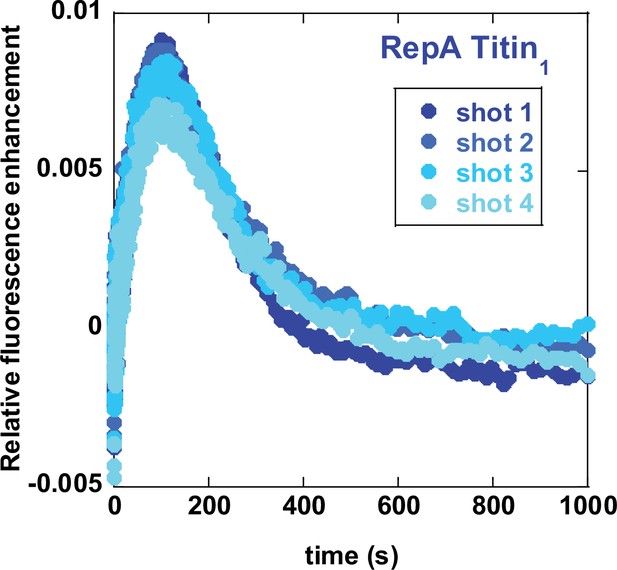
Reproducibility using sequential mixing strategy.
Successive time-courses for RepA-Titin1 were collected using the schematic in Figure 3A. Shots 1–4 indicate time-courses collected in succession.
-
Figure 3—figure supplement 1—source data 1
The sheet consists of data points for each consecutive shot in a tabulated form for RepA-Titin1 substrate, as shown in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig3-figsupp1-data1-v1.xlsx
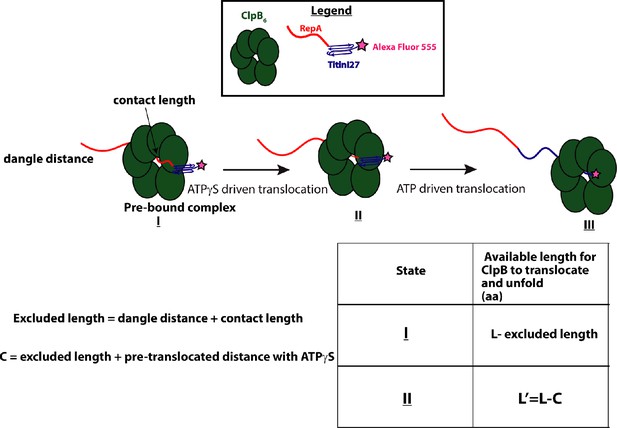
Cartoon representation showing translocation and unfolding by ClpB on RepA-Titin1 from states I through III: (I) Pre-bound complex of ClpB (green) with RepA-Titin1 has dangle distance and contact length unavailable for translocation and unfolding.
(II) After ATPγS-driven translocation has occurred, the unavailable length is the pre-translocated distance and excluded length. (III) On ATP-driven translocation, ClpB completely unfolds and translocates on RepA-Titin1.
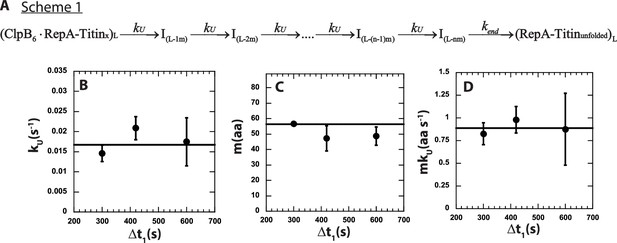
n-Step sequential mechanism for 500:500 μM ATP:ATPγS.
(A) Proposed kinetic scheme for ClpB catalyzed protein unfolding and translocation on RepA-TitinX substrates. Parameters (B) kU, (C) m, and (D) mkU obtained from fitting to Scheme 1 at each Δt1 are shown in solid black circles. The black solid line represents the best weighted fit line to a linear equation with zero slope to yield the average value of the unfolding rate constant, kU = (0.017 ± 0.002) s−1, kinetic step-size, m = (56.5 ± 0.7) aa, and overall rate, mkU = (0.89 ± 0.09) aa s−1.
-
Figure 4—source data 1
The sheet labeled ‘Figure 4B’ consists of data points for the unfolding rate constant, kU as a function of Δt1, represented in Figure 4B.
The error bars for each kU are tabulated as standard deviation. The sheet labeled ‘Figure 4C’ consists of data points for step-size, m as a function of Δt1, represented in Figure 4C. The error bars for each m are tabulated as standard deviation. The sheet labeled ‘Figure 4D’ consists of data points for step-size, mkU as a function of Δt1, represented in Figure 4D. The error bars for each mkU are tabulated as standard deviation.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig4-data1-v1.xlsx
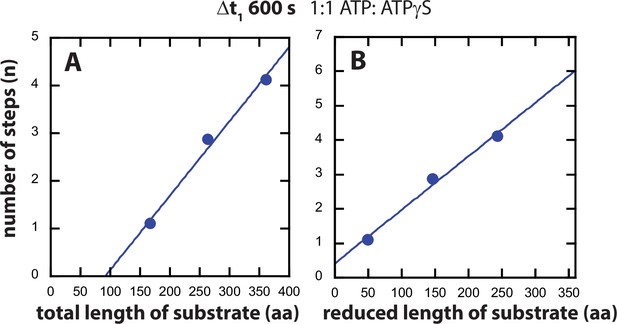
Determination of number of steps, n, from fitting the time-courses represented in Figure 3B to Scheme 1.
The number of steps, n (shown in circles) is plotted as a function of (A) number of steps vs. total length of substrate. Solid line yields a slope = (0.016 ± 0.002) aa−1 and intercept = (−1.4 ± 0.4) steps. (B) Reduced length of substrate calculated by subtracting C = 118 aa from the length. Solid line yields a slope = (0.016 ± 0.002) aa−1 and zero intercept.
-
Figure 4—figure supplement 1—source data 1
The sheet consists of data points for Figure 4—figure supplement 1.
n data points as a function of total length of substrate (Figure 4—figure supplement 1A) and reduced length of substrate (Figure 4—figure supplement 1B) are tabulated here.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig4-figsupp1-data1-v1.xlsx
ClpB catalyzed protein unfolding at approx. 3:1 ATP:ATPγS.
Sequential mixing stopped-flow experiments were carried out as described in Figure 3A with the exception that Syringe 2 contains 600 μM ATPγS. Thus, after both mixing events, the final concentrations are 2 μM ClpB, 50 nM RepA-TitinX, 150 μM ATPγS, 500 μM ATP, and 20 μM α-casein. (A) Representative time-courses from the average of five or more sequentially collected time-courses for RepA-Titin1(blue), RepA-Titin2(purple), and RepA-Titin3(green) at Δt1 = 600 s. The black solid lines represent the best-fit line from fitting to Scheme 1. The fitting parameters obtained are kU = (0.055 ± 0.005) s−1 and m = (58.4 ± 3.8) aa. (B) Total length of substrate as a function of peak time determined for Δt1 = 300, 420, and 600 s as described in Materials and methods. Solid lines represent weighted linear fits yielding (C) slope vs. Δt1 with solid line representing the weighted average of (4.3 ± 0.2) aa s−1 and (D) the intercept vs. Δt1 fit to a linear equation with a slope = (0.05 ± 0.05) aa s−1 and intercept of (67 ± 23) aa. All data points and error bars represent the average and standard deviation determined from three replicates.
-
Figure 4—figure supplement 2—source data 1
The sheet labeled as ‘Figure A’ consists of data points in a tabulated form for each RepA-TitinX substrate, as shown in Figure 4—figure supplement 2A.
The error bars for each relative fluorescence enhancement data point are tabulated here as standard deviation. The data points corresponding to the fits shown in black in Figure 4—figure supplement 2A are tabulated after ‘fit data to Scheme 1 for each RepA-TitinX substrate. The sheet labeled ‘Supp Figure B’ are data points for total length vs. peak time plot shown in Figure 4—figure supplement 2B at each Δt1. The sheet labeled ‘Supp Figure C’ has data points for the rate of translocation as a function of Δt1 shown in Figure 4—figure supplement 2C. The error bars represented in Figure 4—figure supplement 2C are tabulated as the standard deviation in this sheet for each corresponding rate of translocation. The sheet labeled ‘Figure D’ consists of data points for pre-translocated distance as a function of Δt1, as shown in Figure 4—figure supplement 2D. The error bars represented in Figure 4—figure supplement 2D are tabulated as the standard deviation in this sheet for each corresponding pre-translocated distance.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig4-figsupp2-data1-v1.xlsx
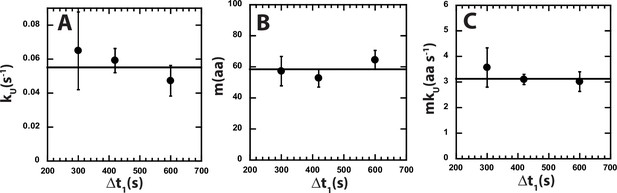
Parameters from fitting to Scheme 1 for approx. 3:1 ATP: ATPγS.
(A) kU, (B) m, and (C) mkU obtained from fitting to Scheme 1 at each Δt1 are shown in solid black circles. The black solid line represents the weighted average value of kU = (0.055 ± 0.005) s−1, m = (58.4 ± 3.8) aa, and mkU = (3.1 ± 0.2) aa s−1. All data points and error bars represent the average and standard deviation determined from three replicates.
-
Figure 4—figure supplement 3—source data 1
The sheet labeled ‘Figure A’ consists of data points for the unfolding rate constant, kU as a function of Δt1, represented in Figure 4—figure supplement 3A.
The error bars for each kU are tabulated as standard deviation. The sheet labeled ‘Figure B’ consists of data points for step-size, m as a function of Δt1, represented in Figure 4—figure supplement 3B. The error bars for each m are tabulated as standard deviation. The sheet labeled ‘Figure C’ consists of data points for step-size, mkU as a function of Δt1, represented in Figure 4—figure supplement 3C. The error bars for each mkU are tabulated as standard deviation.
- https://cdn.elifesciences.org/articles/99052/elife-99052-fig4-figsupp3-data1-v1.xlsx
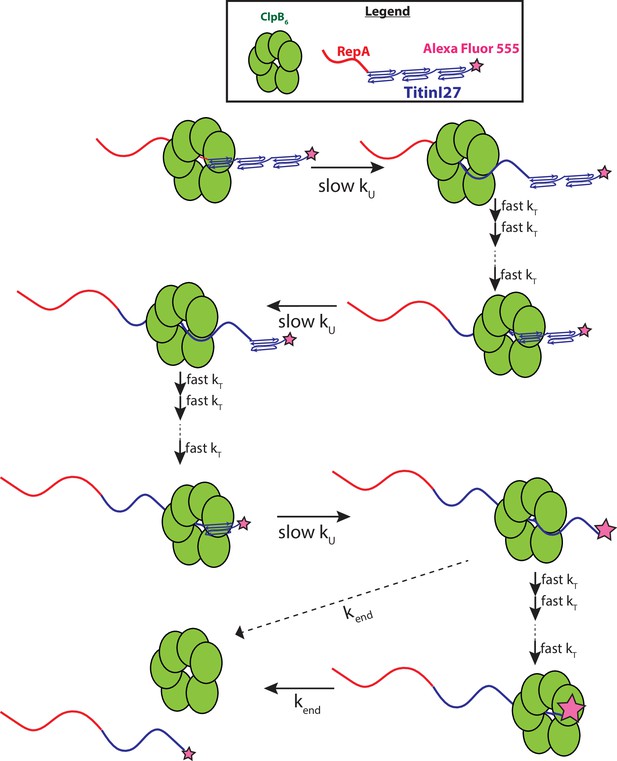
Proposed mechanism of ClpB catalyzed protein unfolding and translocation.
In the presence of ATPγS, hexameric ClpB binds to the unfolded 70 amino acid long RepA sequence. Upon mixing with ATP, ClpB unfolds ~60 amino acids with rate constant, kU, shown as complete collapse of the first Titin I27 domain. ClpB then proceeds through multiple fast translocation steps with rate constant kT before arrival at the next folded Titin I27 domain and the process repeats. After complete unfolding of the final Titin I27 domain, ClpB may dissociate before arrival at the C-terminus, or rapidly translocate to the C-terminus followed by slow dissociation with dissociation rate constant kend. We expect protein-induced fluorescence enhancement (PIFE) to occur at the last unfolding step and/or at the last translocation step; the relative intensities of AF555 represented by the size of the pink star.
Tables
Parameters obtained from model-independent analysis on experiments presented in Figure 3 and Figure 4—figure supplement 2.
| Parameters from model-independent analysis | 500 μM [ATPγS] and 500 μM [ATP] | 150 μM [ATPγS] and 500 μM [ATP] |
|---|---|---|
| Rate of translocation with ATPγS and ATP (aa s−1) | 0.9 ± 0.1 | 4.3 ± 0.2 |
| Excluded length (aa) | 48 ± 17 | 67 ± 23 |
| Rate of translocation with ATPγS (aa s−1) | 0.09 ± 0.06 | 0.05 ± 0.05 |
-
Error represent the standard deviation determined from three replicates.
Parameters obtained from global fitting the time-courses obtained from experiments in Figure 3 and Figure 4—figure supplement 2 to Scheme 1 in Figure 4A.
| Parameters from model-dependent analysis | 500 μM [ATPγS] and 500 μM [ATP] | 150 μM [ATPγS] and 500 μM [ATP] |
|---|---|---|
| m (aa) | 56.5 ± 0.7 | 58 ± 4 |
| kU (s–1) | 0.017 ± 0.002 | 0.055 ± 0.005 |
| mkU (aa s−1) | 0.89 ± 0.09 | 3.1 ± 0.2 |
-
Error represent the standard deviation determined from three replicates.





