A VgrG2b fragment cleaved by caspase-11/4 promotes Pseudomonas aeruginosa infection through suppressing the NLRP3 inflammasome
Figures
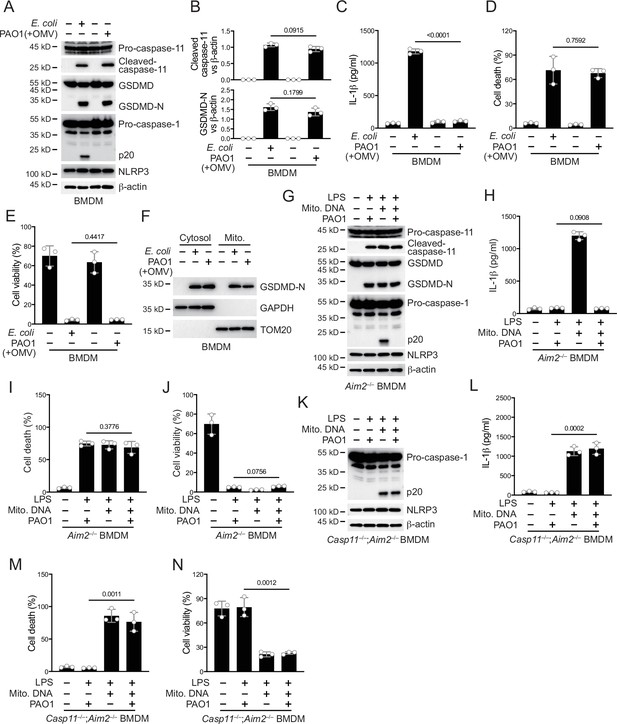
P. aeruginosa infection suppresses NLRP3 inflammasome.
(A–E) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4 and incubated with E. coli or P. aeruginosa (ΔRetS PAO1) at a multiplicity of infection (MOI) of 30 and 20 μg/ml outer membrane vesicles (OMVs) for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection (A). Band intensities of cleaved caspase-11 (top) and GSDMD (bottom) were quantified and compared to that of β-actin (B). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (C). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants 16 hr post infection (D). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (E). (F) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4 and incubated with E. coli or ΔRetS PAO1 at an MOI of 30 and 20 μg/ml OMVs for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were subjected to cellular component fractionation and immunoblotted as indicated 16 hr post infection. (G–J) Aim2–/– BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml lipopolysaccharide (LPS) and 2 μg/ml extracted BMDM mitochondrial DNA using DOTAP with or without the incubation of ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted 16 hr post infection (G). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (H). Cytotoxicity was determined by LDH release assay in cell culture supernatants 16 hr post infection (I). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (J). (K–N) Casp11–/–;Aim2–/– BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS and 2 μg/ml extracted BMDM mitochondrial DNA using DOTAP with or without the incubation of ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted 16 hr post infection (K). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (L). Cytotoxicity was determined by LDH release assay in cell culture supernatants 16 hr post infection (M). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (N). Data were shown as means ± SD. For C–E, H–J, L–N, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 1—source data 1
File containing labeled original western blots for Figure 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig1-data1-v1.zip
-
Figure 1—source data 2
Original gel image files for western blot analysis displayed in Figure 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig1-data2-v1.zip
-
Figure 1—source data 3
Original source data for graphs displayed in Figure 1, Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig1-data3-v1.xlsx
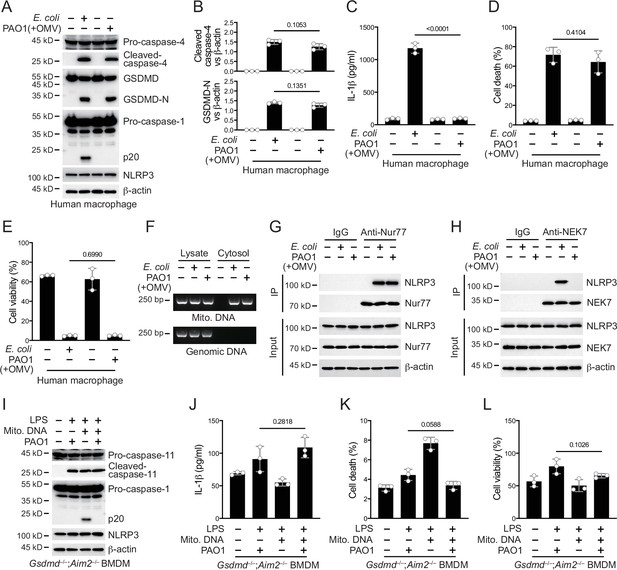
P. aeruginosa infection suppresses NLRP3 inflammasome in human macrophages.
(A–E) Human macrophages were primed overnight with 1 μg/ml Pam3CSK4 and incubated with E. coli or P. aeruginosa (ΔRetS PAO1) at a multiplicity of infection (MOI) of 30 and 20 μg/ml outer membrane vesicles (OMVs) for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted 16 hr post infection (A). Band intensities of cleaved caspase-4 (top) and GSDMD (bottom) were quantified and compared to that of β-actin (B). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (C). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants 16 hr post infection (D). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (E). (F) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4 and incubated with E. coli or ΔRetS PAO1 at an MOI of 30 and 20 μg/ml OMVs for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were subjected to cellular component fractionation and PCR analysis of mitochondrial or genomic DNAs 16 hr post infection. (G, H) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4 and incubated with E. coli or ΔRetS PAO1 at an MOI of 30 and 20 μg/ml OMVs for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoprecipitated with a control IgG or antibody against Nur77 (G) or NEK7 (H) 16 hr post infection. Precipitates were immunoblotted as indicated. (I–L) Gsdmd–/–;Aim2–/– BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS and 2 μg/ml extracted BMDM mitochondrial DNA using DOTAP with or without the incubation of ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted 16 hr post infection (I). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (J). Cytotoxicity was determined by LDH release assay in cell culture supernatants 16 hr post infection (K). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (L). Data were shown as means ± SD. For C–E, J–L, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 1—figure supplement 1—source data 1
File containing labeled original western blots for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Original gel image files for western blot analysis displayed in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig1-figsupp1-data2-v1.zip
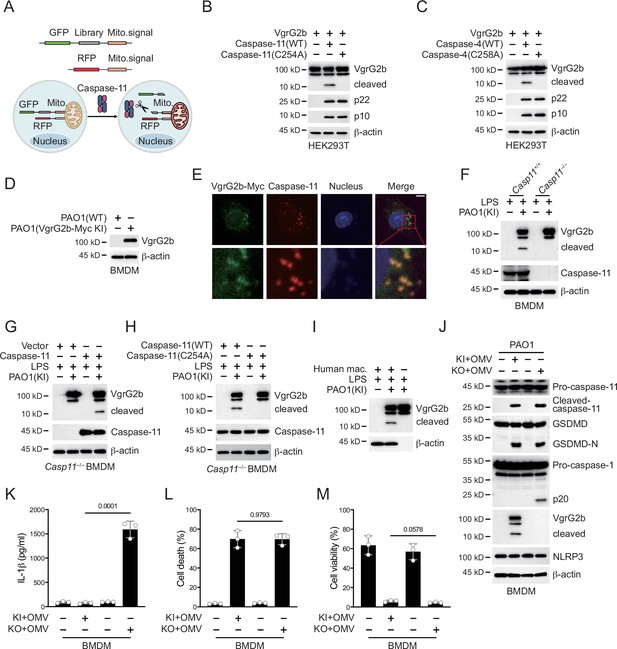
P. aeruginosa VgrG2b is cleaved by caspase-11.
(A) Screening strategy for identifying caspase-11 substrates in P. aeruginosa. Briefly, P. aeruginosa cDNAs were cloned into GFP vectors containing a mitochondrial localization signal. RFP vectors with a mitochondrial localization signal were used as transfection controls. HEK293T cells were co-transfected with these vectors and caspase-11. Cells losing GFP signals on mitochondria were sequenced. Plasmids encoding VgrG2b and mouse caspase-11 p22/p10 (B) or human caspase-4 p22/p10 (C) were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (D) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4 and incubated with ΔRetS PAO1 or VgrG2b-Myc knockin ΔRetS PAO1 at a multiplicity of infection (MOI) of 30 and 20 μg/ml outer membrane vesicles (OMVs) for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection. (E) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4 and incubated with VgrG2b-Myc knockin ΔRetS PAO1 at an MOI of 30 and 20 μg/ml OMVs for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were fixed and stained with antibodies against Myc or caspase-11 16 hr post infection. Nucleus was stained with DAPI. Scale bar, 5 μm. (F) Casp11+/+ and Casp11–/– BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml lipopolysaccharide (LPS) using DOTAP with or without the incubation of VgrG2b-Myc knockin ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted 16 hr post infection. (G) Casp11–/– BMDM cells were infected with lentiviruses encoding a control vector of caspase-11. Cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the incubation of VgrG2b-Myc knockin ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection. (H) Casp11–/– BMDM cells were infected with lentiviruses encoding wild-type or mutant caspase-11. Cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the incubation of VgrG2b-Myc knockin ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection. (I) Human macrophages were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the incubation of VgrG2b-Myc knockin ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection. (J–M) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by incubation of VgrG2b-Myc knockin (KI) or knockout (KO) ΔRetS PAO1 at an MOI of 30 and 20 μg/ml OMVs for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection (J). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (K). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants 16 hr post infection (L). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (M). Data were shown as means ± SD. For K–M, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 2—source data 1
File containing labeled original western blots for Figure 2.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig2-data1-v1.zip
-
Figure 2—source data 2
Original gel image files for western blot analysis displayed in Figure 2.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig2-data2-v1.zip
-
Figure 2—source data 3
Original source data for graphs displayed in Figure 2, Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig2-data3-v1.xlsx
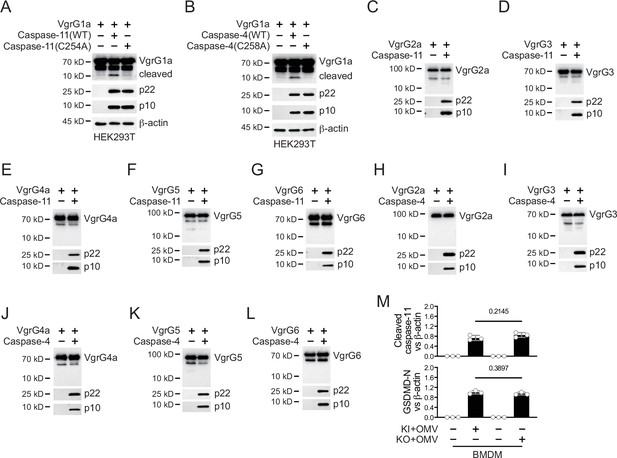
Caspase-11 cleaves limited VgrG family members.
Plasmids encoding VgrG1a and mouse caspase-11 p22/p10 (A) or human caspase-4 p22/p10 (B) were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (C–L) Plasmids encoding the indicated VgrG family members and mouse caspase-11 p22/p10 or human caspase-4 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (M) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by incubation of VgrG2b-Myc knockin (KI) or knockout (KO) ΔRetS PAO1 at a multiplicity of infection (MOI) of 30 and 20 μg/ml outer membrane vesicles (OMVs) for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted 16 hr post infection. Band intensities of cleaved caspase-11 (top) and GSDMD (bottom) were quantified and compared to that of β-actin. Data were shown as means ± SD. Experiments were repeated three times with similar results.
-
Figure 2—figure supplement 1—source data 1
File containing labeled original western blots for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Original gel image files for western blot analysis displayed in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig2-figsupp1-data2-v1.zip
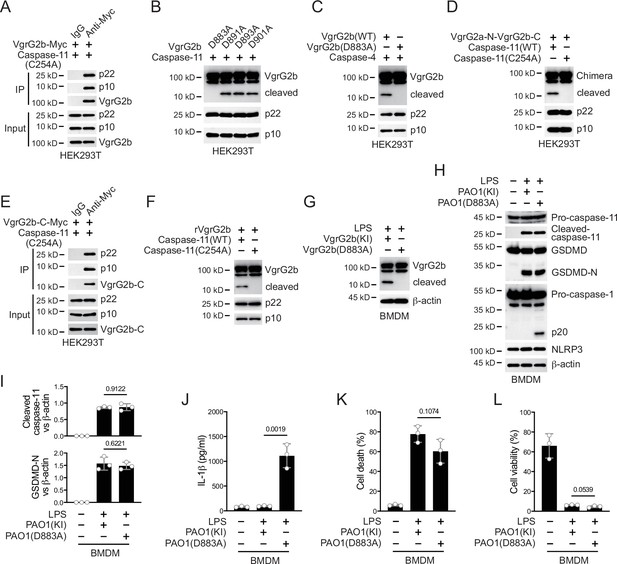
Caspase-11 cleaves VgrG2b at D883.
(A) Plasmids encoding VgrG2b and mutant caspase-11 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (B) Plasmids encoding caspase-11 p22/p10 and VgrG2b mutants were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (C) Plasmids encoding caspase-4 p22/p10 and VgrG2b variants were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (D) Plasmids encoding VgrG2a-N-VgrG2b-C chimera and caspase-11 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (E) Plasmids encoding VgrG2b C-terminus and mutant caspase-11 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (F) Recombinant VgrG2b were incubated with caspase-11 p22/p10 for 4 hr, followed by immunoblotting with antibodies against the indicated proteins. (G) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml lipopolysaccharide (LPS) using DOTAP with or without the incubation of VgrG2b wild-type (KI) or mutant (D883A) knockin ΔRetS PAO1 at a multiplicity of infection (MOI) of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted 16 hr post infection. (H–L) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the incubation of VgrG2b wild-type (KI) or mutant (D883A) knockin ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection (H). Band intensities of cleaved caspase-11 (top) and GSDMD (bottom) were quantified and compared to that of β-actin (I). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (J). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants 16 hr post infection (K). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (L). Data were shown as means ± SD. For J–L, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 3—source data 1
File containing labeled original western blots for Figure 3.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig3-data1-v1.zip
-
Figure 3—source data 2
Original gel image files for western blot analysis displayed in Figure 3.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig3-data2-v1.zip
-
Figure 3—source data 3
Original source data for graphs displayed in Figure 3, Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig3-data3-v1.xlsx
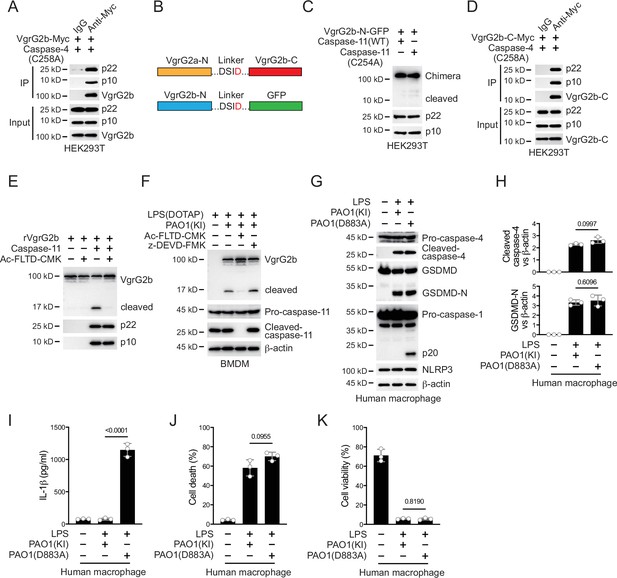
Caspase-11 cleaves VgrG2b during P. aeruginosa infection.
(A) Plasmids encoding VgrG2b and mutant caspase-4 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (B) Scheme for VgrG2b chimeras. (C) Plasmids encoding VgrG2b-N-GFP chimera and caspase-11 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (D) Plasmids encoding VgrG2b-C and mutant caspase-4 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (E) Recombinant VgrG2b was incubated with caspase-11 p22/p10 subunits with or without the presence of 10 μM Ac-FLTD-CMK, followed by immunoblotting with antibodies against the indicated proteins. (F) BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml lipopolysaccharide (LPS) using DOTAP with or without the incubation of VgrG2b-Myc knockin (KI) ΔRetS PAO1 at a multiplicity of infection (MOI) of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin and 10 μM Ac-FLTD-CMK or 10 μM z-DEVD-FMK. Cells were lysed and immunoblotted 16 hr post infection. (G–K) Human macrophages were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the incubation of VgrG2b (KI) or mutant (D883A) knockin ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection (G). Band intensities of cleaved caspase-4 (top) and GSDMD (bottom) were quantified and compared to that of β-actin (H). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (I). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants 16 hr post infection (J). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (K). Data were shown as means ± SD. For I–K, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 3—figure supplement 1—source data 1
File containing labeled original western blots for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Original gel image files for western blot analysis displayed in Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig3-figsupp1-data2-v1.zip
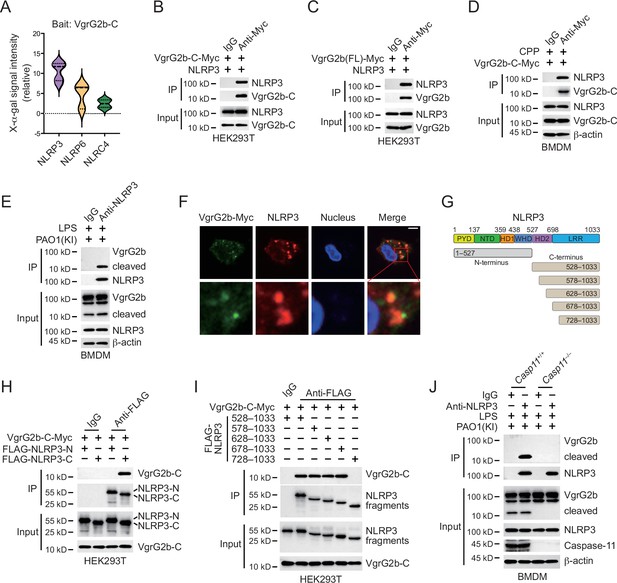
VgrG2b C-terminus interacts with NLRP3.
(A) Yeast two-hybrid screening was performed using VgrG2b C-terminus as bait and a mouse bone marrow cDNA library was screened. The interaction strength between bait and preys were visualized by X-α-gal assays. Plasmids encoding VgrG2b C-terminus (B) or full-length (C) and NLRP3 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (D) VgrG2b C-terminus proteins were introduced into BMDM cells with the help of cell-penetrating peptides (CPP) for 6 hr. Cells were lysed and immunoprecipitated with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (E) BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml lipopolysaccharide (LPS) using DOTAP with the incubation of VgrG2b knockin (KI) ΔRetS PAO1 at a multiplicity of infection (MOI) of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoprecipitated with a control IgG or antibody against NLRP3 16 hr post infection. Precipitates were immunoblotted as indicated. (F) BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with the incubation of VgrG2b knockin (KI) ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were fixed and stained with antibodies against Myc and NLRP3 16 hr post infection. Nucleus was stained with DAPI. Scale bar, 5 μm. (G) Scheme for NLRP3 truncations. (H, I) Plasmids encoding VgrG2b C-terminus and NLRP3 truncations were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against FLAG. Precipitates were immunoblotted as indicated. (J) Casp11+/+ and Casp11–/– BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with the incubation of VgrG2b-Myc knockin (KI) ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoprecipitated with a control IgG or antibody against NLRP3 16 hr post infection. Precipitates were immunoblotted as indicated. Data were shown as means ± SD. Experiments were repeated three times with similar results.
-
Figure 4—source data 1
File containing labeled original western blots for Figure 4.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig4-data1-v1.zip
-
Figure 4—source data 2
Original gel image files for western blot analysis displayed in Figure 4.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig4-data2-v1.zip
-
Figure 4—source data 3
Original source data for graphs displayed in Figure 4.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig4-data3-v1.xlsx
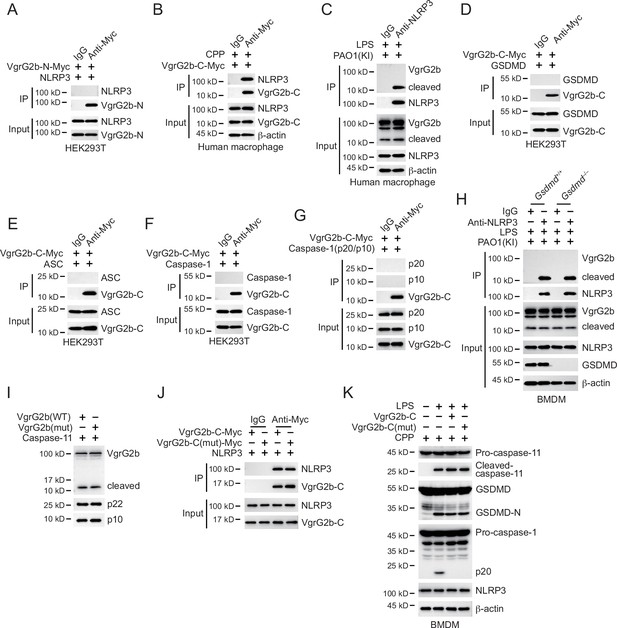
Cleaved VgrG2b fragment binds to NLRP3.
(A) Plasmids encoding VgrG2b N-terminus and NLRP3 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (B) VgrG2b C-terminus proteins were introduced into human macrophage cells with the help of cell-penetrating peptides (CPP) for 6 hr. Cells were lysed and immunoprecipitated with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (C) Human macrophage cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml lipopolysaccharide (LPS) using DOTAP with the incubation of VgrG2b knockin (KI) ΔRetS PAO1 at a multiplicity of infection (MOI) of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoprecipitated with a control IgG or antibody against NLRP3 16 hr post infection. Precipitates were immunoblotted as indicated. Plasmids encoding VgrG2b C-terminus and GSDMD (D), ASC (E), caspase-1 (F), and caspase-1 p20/p10 (G) were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (H) Gsdmd+/+ and Gsdmd–/– BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with the incubation of VgrG2b-Myc knockin (KI) ΔRetS PAO1 at an MOI of 30 for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoprecipitated with a control IgG or antibody against NLRP3 16 hr post infection. (I) Plasmids encoding wild-type VgrG2b, mutant VgrG2b (H935A;H936A;H939A;E983A), and caspase-11 p22/p10 were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (J) Plasmids encoding wild-type VgrG2b C-terminus, mutant VgrG2b C-terminus (H935A;H936A;H939A;E983A) and NLRP3 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (K) BMDM cells were primed overnight with 1 μg/ml Pam3CSK4. Recombinant wild-type VgrG2b C-terminus and mutant VgrG2b C-terminus (H935A;H936A;H939A;E983A) were introduced into BMDM cells with the help of cell-penetrating peptides (CPP) for 6 hr, followed by transfection of 2 μg/ml LPS using DOTAP. Cells were immunoblotted with antibodies against the indicated proteins. Precipitates were immunoblotted as indicated. Data were shown as means ± SD. Experiments were repeated three times with similar results.
-
Figure 4—figure supplement 1—source data 1
File containing labeled original western blots for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original gel image files for western blot analysis displayed in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig4-figsupp1-data2-v1.zip
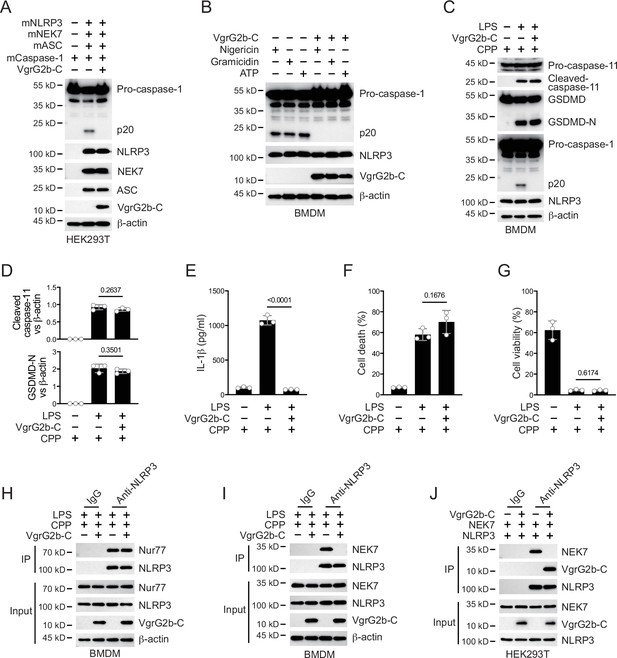
VgrG2b C-terminus suppresses the NLRP3 inflammasome.
(A) Plasmids encoding VgrG2b C-terminus and the indicated murine proteins were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (B) Wild-type BMDM cells were infected with lentiviruses encoding a control vector or VgrG2b C-terminus and primed with 1 μg/ml lipopolysaccharide (LPS) for 3 hr, followed by stimulation of 10 μM nigericin for 30 min, 0.5 μM gramicidin for 1 hr and 2 mM ATP for 30 min. Cells were lysed and immunoblotted as indicated. (C–G) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the transfection of VgrG2b C-terminus proteins using cell-penetrating peptides (CPP) for 2 hr. Cells were then supplemented with fresh medium. Cells were lysed and immunoblotted 16 hr post transfection (C). Band intensities of cleaved caspase-11 (top) and GSDMD (bottom) were quantified and compared to that of β-actin (D). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (E). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants 16 hr post infection (F). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (G). (H, I) Wild-type BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the transfection of VgrG2b C-terminus proteins using cell-penetrating peptides (CPP) for 2 hr. Cells were then supplemented with fresh medium. Cells were lysed and immunoprecipitated with a control IgG or antibody against NLRP3 16 hr post transfection. Precipitates were immunoblotted with antibody against Nur77 (H) or NEK7 (I). (J) Plasmids encoding VgrG2b C-terminus, NEK7 and NLRP3 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against NLRP3. Precipitates were immunoblotted as indicated. Data were shown as means ± SD. For E–G, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 5—source data 1
File containing labeled original western blots for Figure 5.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig5-data1-v1.zip
-
Figure 5—source data 2
Original gel image files for western blot analysis displayed in Figure 5.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig5-data2-v1.zip
-
Figure 5—source data 3
Original source data for graphs displayed in Figure 5, Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig5-data3-v1.xlsx
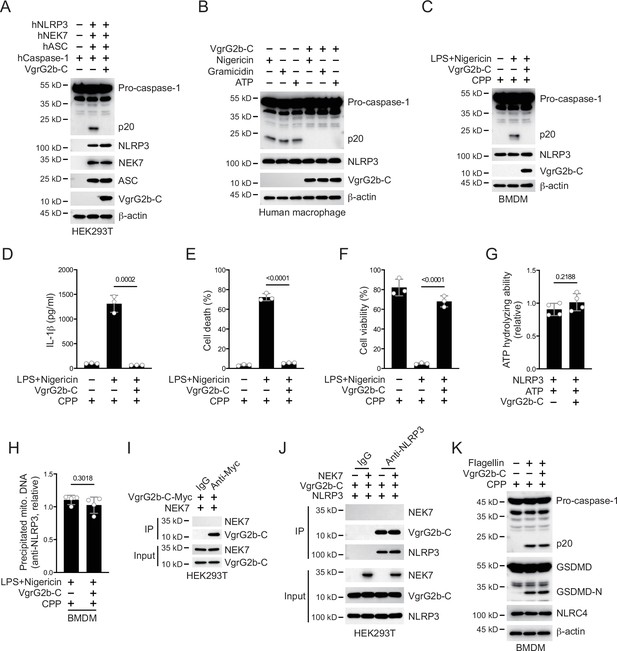
VgrG2b C-terminus inhibits NLRP3 inflammasome activation in human cells.
(A) Plasmids encoding VgrG2b C-terminus and the indicated human proteins were co-transfected into HEK293T cells for 24 hr, followed by immunoblotting with antibodies against the indicated proteins. (B) Human macrophage cells were infected with lentiviruses encoding a control vector or VgrG2b C-terminus and primed with 1 μg/ml lipopolysaccharide (LPS) for 3 hr, followed by stimulation of 10 μM nigericin for 30 min, 0.5 μM gramicidin for 1 hr and 2 mM ATP for 30 min. Cells were lysed and immunoblotted as indicated. (C–F) Wild-type BMDM cells were primed with 1 μg/ml LPS for 3 hr, followed by stimulation of 10 μM nigericin with or without the transfection of VgrG2b C-terminus proteins using cell-penetrating peptides (CPP) for 30 min. Cells were lysed and immunoblotted (C). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels (D). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants (E). Cell viability was determined by an ATP quantification assay in cell pellets (F). (G) FLAG-NLRP3 was purified from HEK293T cells using anti-FLAG-conjugated beads. NLRP3-containing beads were incubated with 100 μCi[γ-32P]ATP in the presence of 100 ng/ml VgrG2b C-terminus. Beads were washed with PBS and the associated radioactive ATP was determined by liquid scintillation. (H) Wild-type BMDM cells were primed with 1 μg/ml LPS for 3 hr, followed by stimulation of 10 μM nigericin with or without the transfection of VgrG2b C-terminus proteins using cell-penetrating peptides (CPP) for 30 min. Cells were lysed and immunoprecipitated with antibody against NLRP3. The amount of mitochondrial DNA in the precipitates was examined through quantitative PCR. (I) Plasmids encoding VgrG2b C-terminus and NEK7 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against Myc. Precipitates were immunoblotted as indicated. (J) Plasmids encoding VgrG2b C-terminus, NEK7 and NLRP3 were co-transfected into HEK293T cells for 24 hr, followed by immunoprecipitation with a control IgG or antibody against NLRP3. Precipitates were immunoblotted as indicated. (K) Recombinant VgrG2b C-terminus proteins were introduced into BMDM cells with the help of cell-penetrating peptides (CPP) for 6 hr. Cells were then transfected with 4 μg/ml flagellin with the help of lipofectin for 4 hr. Cells were lysed and immunoprecipitated with antibodies against the indicated proteins. Data were shown as means ± SD. For D–F, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 5—figure supplement 1—source data 1
File containing labeled original western blots for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Original gel image files for western blot analysis displayed in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig5-figsupp1-data2-v1.zip
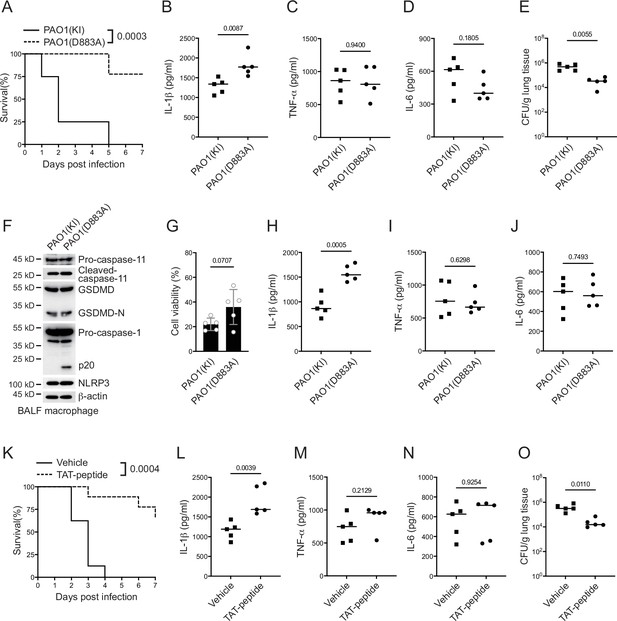
P. aeruginosa suppresses NLRP3 inflammasome in vivo.
(A–E) Wild-type mice were intraperitoneally challenged with poly(I:C) at a dose of 2 mg/kg body weight for 6 hr and then intranasally infected with 1 × 109 cfu of VgrG2b (KI) or mutant (D883A) knockin ΔRetS PAO1, followed by survival calculation at the indicated days (A). Serum protein levels of IL-1β (B), TNF-α (C), and IL-6 (D) were examined through ELISA assays 24 hr post infection. Bacterial load was examined in mouse lung tissues 4 days post infection (E). (F–J) Wild-type mice were intraperitoneally challenged with poly(I:C) at a dose of 2 mg/kg body weight for 6 hr and then intranasally infected with 1 × 109 cfu of VgrG2b (KI) or mutant (D883A) knockin ΔRetS PAO1. Bronchoalveolar lavage fluids (BALF) were collected 16 hr post infection. Cells were immunoblotted as indicated (F). Cell viability was also determined by an ATP quantification assay in cell pellets (G). Protein levels of IL-1β (H), TNF-α (I), and IL-6 (J) in BALF supernatants were examined through ELISA assays. (K–O) Wild-type mice were intraperitoneally challenged with poly(I:C) at a dose of 2 mg/kg body weight for 6 hr and then intranasally infected with 1 × 109 cfu of ΔRetS PAO1 and administrated with 20 μg TAT sequence containing VgrG2b C-terminus peptide, followed by survival calculation at the indicated days (K). Serum protein levels of IL-1β (L), TNF-α (M), and IL-6 (N) were examined through ELISA assays 24 hr post infection. Bacterial load was examined in mouse lung tissues 4 days post infection (O). Data were shown as means ± SD. Experiments were repeated three times with similar results.
-
Figure 6—source data 1
File containing labeled original western blots for Figure 6.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig6-data1-v1.zip
-
Figure 6—source data 2
Original gel image files for western blot analysis displayed in Figure 6.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig6-data2-v1.zip
-
Figure 6—source data 3
Original source data for graphs displayed in Figure 6, Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig6-data3-v1.xlsx
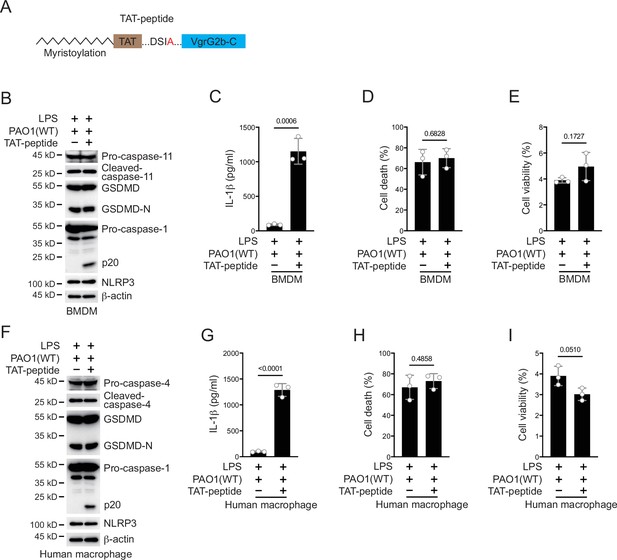
Activation of NLRP3 inflammasome ameliorates P. aeruginosa infection.
(A) Scheme for design of a VgrG2b C-terminus peptide. VgrG2b C-terminus was added with an N-terminal TAT sequence, followed by myristoylation. (B–E) BMDM cells were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml lipopolysaccharide (LPS) using DOTAP with or without the incubation of ΔRetS PAO1 at a multiplicity of infection (MOI) of 30 and 2 μg/ml TAT-peptide for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection (B). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (C). Cytotoxicity was determined by lactate dehydrogenase (LDH) release assay in cell culture supernatants 16 hr post infection (D). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (E). (F–I) Human macrophages were primed overnight with 1 μg/ml Pam3CSK4, followed by transfection of 2 μg/ml LPS using DOTAP with or without the incubation of ΔRetS PAO1 at an MOI of 30 and 2 μg/ml TAT-peptide for 2 hr. Cells were then supplemented with fresh medium containing 100 μg/ml Gentamycin. Cells were lysed and immunoblotted as indicated 16 hr post infection (F). Cell culture supernatants were collected for an ELISA assay to determine the secreted IL-1β protein levels 16 hr post infection (G). Cytotoxicity was determined by LDH release assay in cell culture supernatants 16 hr post infection (H). Cell viability was determined by an ATP quantification assay in cell pellets 16 hr post infection (I). Data were shown as means ± SD. For C–E, G–I, data of three independent experiments were calculated. Experiments were repeated three times with similar results.
-
Figure 6—figure supplement 1—source data 1
File containing labeled original western blots for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Original gel image files for western blot analysis displayed in Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/99939/elife-99939-fig6-figsupp1-data2-v1.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Casp11–/– mice | Cyagen Biosciences (Jiangsu, China) | S-KO-01332 | |
| Strain, strain background (Mus musculus) | Gsdmd–/– | Cyagen Biosciences (Jiangsu, China) | S-KO-12963 | |
| Strain, strain background (Mus musculus) | Aim2–/– | Cyagen Biosciences (Jiangsu, China) | S-KO-09889 | |
| Strain, strain background (Escherichia coli) | E. coli | China General Microbiological Culture Collection Center | ATCC11775 | |
| Strain, strain background (Pseudomonas aeruginosa) | PAO1 | Institute of Microbiology, Chinese Academy of Sciences | CGMCC1.12483 | |
| Antibody | anti-NLRP3 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 15101 | (1:1000) |
| Antibody | anti-IL-1β (Rabbit monoclonal) | Cell Signaling Technology | Cat# 12703 | (1:1000) |
| Antibody | anti-IL-1β (Rabbit monoclonal) | Cell Signaling Technology | Cat# 12426 | (1:1000) |
| Antibody | anti-cleaved IL-1β (Rabbit monoclonal) | Cell Signaling Technology | Cat# 83186 | (1:1000) |
| Antibody | anti-cleaved IL-1β (Rabbit monoclonal) | Cell Signaling Technology | Cat# 63124 | (1:1000) |
| Antibody | anti-GSDMD (Rabbit polyclonal) | Cell Signaling Technology | Cat# 93709 | (1:1000) |
| Antibody | anti- caspase-1 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 3866 | (1:1000) |
| Antibody | anti-caspase-1 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 24232 | (1:1000) |
| Antibody | anti-caspase-1 (Rabbit polyclonal) | Cell Signaling Technology | Cat# 2225 | (1:1000) |
| Antibody | anti-ASC (Rabbit monoclonal) | Cell Signaling Technology | Cat# 13833 | (1:1000) |
| Antibody | anti-ASC (Rabbit monoclonal) | Cell Signaling Technology | Cat# 67824 | (1:1000) |
| Antibody | anti-caspase-1 (Rabbit monoclonal) | Abcam | Cat# ab207802 | (1:1000) |
| Antibody | anti-NEK7 (Rabbit monoclonal) | Abcam | Cat# ab133514 | (1:10,000) |
| Antibody | anti-Nur77 (Rabbit polyclonal) | Abcam | Cat# ab153914 | (1:1000) |
| Antibody | anti-GSDMD (Rabbit monoclonal) | Abcam | Cat# ab215203 | (1:1000) |
| Antibody | anti-caspase-11 (Rabbit monoclonal) | Abcam | Cat# ab180673 | (1:1000) |
| Antibody | anti-caspase-11 (Rat monoclonal) | Cell Signaling Technology | Cat# 14340 | (1:1000) |
| Antibody | anti-caspase-11 (Rat monoclonal) | Invitrogen | Cat# 14-9935-82 | (1:1000) |
| Antibody | anti-caspase-11 (Mouse monoclonal) | MBL Beijing Biotech | Cat# M029-3 | (1:1000) |
| Antibody | anti-6xHis tag (Mouse monoclonal) | Invitrogen | Cat# MA121315 | (1:1000) |
| Antibody | anti-FLAG tag (Mouse monoclonal) | Sigma-Aldrich | Cat# F3165 | (1:1000) |
| Antibody | anti-β-actin (Mouse monoclonal) | Sigma-Aldrich | Cat# A1978 | (1:2000) |
| Antibody | anti-c-Myc (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-40 | (1:1000) |
| Antibody | anti-Tom20 (Rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-11415 | (1:500) |
| Antibody | anti-GAPDH (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-32233 | (1:1000) |
| Antibody | anti-HA tag (Rabbit polyclonal) | Huaxingbio (Beijing) | Cat# HX1820 | (1:5000) |
| Antibody | HRP-conjugated anti-mouse IgG (Goat polyclonal) | Proteintech | Cat# SA00001-1 | (1:5000) |
| Antibody | HRP-conjugated goat anti-rabbit IgG (Goat polyclonal) | Proteintech | Cat# SA00001-2 | (1:5000) |
| Sequence-based reagent | RetS-up-F | This paper | PCR primers | ctcggtacccggggatccccgc cgtgcgcgacatgctcgccggcaa |
| Sequence-based reagent | RetS-up-R | This paper | PCR primers | agcacgtcgctgccc ggcgaagtcccttcg |
| Sequence-based reagent | RetS-down-F | This paper | PCR primers | ccttcgaagggacttcgccg ggcagcgacgtgctccgg |
| Sequence-based reagent | RetS-down-R | This paper | PCR primers | cgttgtaaaacgacggccagtgcca agcttcgagggtcaggcaggcgag |
| Sequence-based reagent | KO-VgrG2b-up-F | This paper | PCR primers | attcgagctcggtacccggggatcc cttgatggaaaagagtttcaagacc |
| Sequence-based reagent | KO-VgrG2b-up-R | This paper | PCR primers | tctcgaggaaataatctcgaacg ataggctcgcagagcgcttcttccagt |
| Sequence-based reagent | KO-VgrG2b-down-F | This paper | PCR primers | actggaagaagcgctctgcgag cctatcgttcgagattatttcctcga |
| Sequence-based reagent | KO-VgrG2b-down-R | This paper | PCR primers | taaaacgacggccagtgccaagc ttgacgacgtcggggttctctgcctt |
| Sequence-based reagent | KI-Myc-VgrG2b-up-F | This paper | PCR primers | tcgagctcggtacccggggatccgag cacatcaccctgatgtgcggcggcgcct |
| Sequence-based reagent | KI-Myc-VgrG2b-up-R1 | This paper | PCR primers | ttcgaaccgcgggccctctagactcga gcggtatcccgttgggaagtttttcagt |
| Sequence-based reagent | KI-Myc-VgrG2b-up-R2 | This paper | PCR primers | atcctcttctgagatgagtttttgttc gaaccgcgggccctctagactc |
| Sequence-based reagent | KI-Myc-VgrG2b-down-F | This paper | PCR primers | aaaaactcatctcagaagaggatctgt gaccaatgaaatgcaagaccttgctca |
| Sequence-based reagent | KI-Myc-VgrG2b-down-R | This paper | PCR primers | taaaacgacggccagtgccaagcttc ggccccgccggcgccactggcgaa |
| Sequence-based reagent | VgrG2b-D883A-up-F | This paper | PCR primers | aattcgagctcggtacccggggatcc gagcaccagggcgtggggcacgacg |
| Sequence-based reagent | VgrG2b-D883A-up-R | This paper | PCR primers | aggtcttgcatttcattggtcac agatcctcttctgagatgag |
| Sequence-based reagent | VgrG2b-D883A-down-F | This paper | PCR primers | actcatctcagaagaggatctgtgac caatgaaatgcaagaccttgctca |
| Sequence-based reagent | VgrG2b-D883A-down-R | This paper | PCR primers | taaaacgacggccagtgccaagct tttcgccggccaggcagaattcgacg |
| Commercial assay or kit | RNA extraction kit | Dakewe (Beijing) | Cat# 8034111 | |
| Commercial assay or kit | CellTiter-Glo luminescent cell viability assay kit | Promega | Cat# G7570 | |
| Commercial assay or kit | CytoTox 96 non-radioactive cytotoxicity assay kit | Promega | Cat# G1780 | |
| Chemical compound, drug | LPS | Innochem (Beijing) | Cat# B46894 | |
| Chemical compound, drug | DOTAP | Psaitong (Beijing) | Cat# D10530 | |
| Chemical compound, drug | DOTAP | Roche | Cat# 11202375001 | |
| Chemical compound, drug | Nigericin | Merck Millipore | Cat# 481990 | |
| Chemical compound, drug | Gramicidin | Merck Millipore | Cat# 368020 | |
| Chemical compound, drug | ATP | Merck Millipore | Cat# A6559 | |
| Chemical compound, drug | Poly(I:C) | InvivoGen | Cat# tlrl-picw | |
| Chemical compound, drug | Pam3CSK4 | InvivoGen | Cat# tlrl-pms | |
| Chemical compound, drug | Cell-penetrating peptides | Abcam | Cat# ab142343 | |
| Chemical compound, drug | Cell-penetrating peptides | Santa Cruz | Cat# sc-396807 | |
| Chemical compound, drug | Human M-CSF | PeproTech | Cat# 300-25 | |
| Chemical compound, drug | Murine M-CSF | PeproTech | Cat# 315-02 | |
| Software, algorithm | GraphPad Prism | GraphPad Software, https://www.graphpad.com/ | RRID:SCR_002798 |





