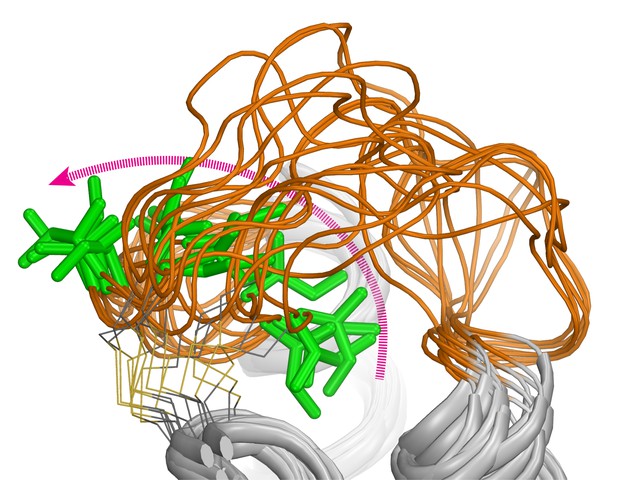
The extracellular loop 2 (orange) of the dopamine D2 receptor is highly dynamic: as one of its elements (green) flips from a buried to an exposed position (magenta arrow), the conformation of the loop changes. Image credits: Lei Shi (CC BY 4.0)
Almost a third of prescribed drugs work by acting on a group of proteins known as GPCRs (short for G-protein coupled receptors), which help to transmit messages across the cell’s outer barrier. The neurotransmitter dopamine, for instance, can act in the brain and body by attaching to dopamine receptors, a sub-family of GPCRs. The binding process changes the three-dimensional structure (or conformation) of the receptor from an inactive to active state, triggering a series of molecular events in the cell.
However, GPCRs do not have a single ‘on’ or ‘off’ state; they can adopt different active shapes depending on the activating molecule they bind to, and this influences the type of molecular cascade that will take place in the cell. Some evidence also shows that classes of GPCRs can have different inactive structures; whether this is also the case for the dopamine D2 and D3 receptors remained unclear. Mapping out inactive conformations of receptors is important for drug discovery, as compounds called antagonists can bind to inactive receptors and interfere with their activation.
Lane et al. proposed that different types of antagonists could prefer specific types of inactive conformations of the dopamine D2 and D3 receptors. Based on the structures of these two receptors, the conformations of D2 bound with the drugs risperidone and eticlopride (two dopamine antagonists) were simulated and compared. The results show that the inactive conformations of D2 were very different when it was bound to eticlopride as opposed to risperidone. In addition D2 and D3 showed a very similar conformation when attached to eticlopride. The two drugs also bound to the inactive receptors in overlapping but different locations. These computational findings, together with experimental validations, suggest that D2 and D3 exist in several inactive states that only allow the binding of specific drugs; these states could also reflect different degrees of inactivation. Overall, the work by Lane et al. contributes to a more refined understanding of the complex conformations of GPCRs, which could be helpful to screen and develop better drugs.




