Scientists have revealed how large molecules are transported into the centre of cells (the cell nucleus) by using large cargo models based on viruses, a new study in eLife reports.
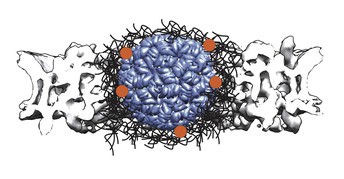
Model of a large molecule (blue, PD ID: 2MS2) bound to multiple transporter proteins (orange dots) that interact with the NPC barrier (black) and help insert the cargo into the NPC (grey structure, EMD-8087). Image credit: Giulia Paci (CC BY 4.0)
The research provides important insights into how large cargoes such as viruses can get inside the nucleus of a cell, which allows them to multiply and infect others.
In the typical mammalian cell, there are more than 2,000 nuclear ‘pores’ that span the surface of the nucleus and allow molecules to enter and exit. These nuclear pores are gatekeepers that restrict the access of molecules over a certain size (approximately five nanometres). However, cargoes that have a nuclear localisation sequence (NLS) on their surface can bind to molecules inside nuclear pores and enter rapidly.
“Nuclear pores are remarkable in the diversity of cargoes they can transport, ranging from the import of proteins and viruses to the export of RNAs and proteins into the cell cytoplasm,” explains senior author Edward Lemke, Professor of Synthetic Biophysics at Johannes Gutenberg University Mainz (JGU) and Adjunct Director of the Institute of Molecular Biology (IMB) in Mainz, Germany. “Despite its high relevance to biology, how large cargoes, such as those greater than 15 nanometres, can be efficiently transported is still an enigma, especially considering the dimensions and structure of the nuclear pore itself.”
To address this gap, the researchers designed a set of large model cargoes based on the structure of viral capsids (the protein shell of a virus). These ranged in size from 17 nanometres to 36 nanometres in diameter. They then engineered the capsid surface with a fluorescent dye and with varying numbers of NLSs. The different labelled capsids were then observed as they entered the nucleus of cells using time-lapse fluorescence microscopy. As more capsids were transported into the nucleus, the fluorescent signal in the nucleus intensified.
They found that in the absence of any NLSs on the capsid surface, all the capsids stayed in the cell cytoplasm and none accumulated in the nucleus. As the number of NLSs on the capsid increased, accumulation of the capsid cargo in the nucleus was more efficient. Most significantly, the larger the capsid, the more NLSs were needed to enable efficient transport into the nucleus.
The team looked at a variety of capsids from different viruses and bacteriophage (viruses that infect bacteria). When they studied the transport of Hepatitis B capsids, the largest cargo used in the experiments, they found that even increasing the number of NLSs on the surface to 240 still did not result in the transport of the capsid into the nucleus. This is in line with previous studies of the Hepatitis B virus which suggest that only the mature infectious virus can move through the nuclear pore into the nucleus.
Having gained insights using their model cargos in real cells, the team then developed a mathematical model to further explore the transport mechanism of large molecules into the nucleus and help establish the main determinants of efficient transport. This model was created in collaboration with the laboratory of Anton Zilman at the Physics Department and Institute for Biomedical Engineering, University of Toronto, Canada.
The team found that their results could be explained using a simple physical model that considers only a minimal number of factors. The model suggests it is harder for large cargoes to enter the nuclear pore and that this cost is overcome by the binding of the capsids with proteins inside the nuclear pore, enabled by the NLS.
“Our simple two-parameter model has recreated the requirements for nuclear transport and revealed key molecular determinants of the transport of large biological cargoes on cells,” concludes first author Giulia Paci, who carried out the study as part of her PhD at the European Molecular Biology Laboratory, Heidelberg, Germany. “Future studies using our model cargoes with high-resolution, timed measurements could provide further insights into the step-by-step movements and binding of large cargoes in the nuclear pore. These studies could also inform the development of more detailed mathematical models to provide further insight on the architecture of the nuclear pore complex."
Media contacts
Emily Packer
eLife
e.packer@elifesciences.org
+441223855373
About
eLife is a non-profit organisation created by funders and led by researchers. Our mission is to accelerate discovery by operating a platform for research communication that encourages and recognises the most responsible behaviours. We work across three major areas: publishing, technology and research culture. We aim to publish work of the highest standards and importance in all areas of biology and medicine, including Cell Biology and Structural Biology and Molecular Biophysics, while exploring creative new ways to improve how research is assessed and published. We also invest in open-source technology innovation to modernise the infrastructure for science publishing and improve online tools for sharing, using and interacting with new results. eLife receives financial support and strategic guidance from the Howard Hughes Medical Institute, the Knut and Alice Wallenberg Foundation, the Max Planck Society and Wellcome. Learn more at https://elifesciences.org/about.
To read the latest Cell Biology research published in eLife, visit https://elifesciences.org/subjects/cell-biology.
And for the latest in Structural Biology and Molecular Biophysics, see https://elifesciences.org/subjects/structural-biology-molecular-biophysics.




